8IG5
 
 | | Crystal structure of SARS main protease in complex with GC376 | | Descriptor: | 3C-like proteinase nsp5, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG6
 
 | | Crystal structure of MERS main protease in complex with GC376 | | Descriptor: | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, ORF1a | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG9
 
 | |
4LDL
 
 | | Structure of beta2 adrenoceptor bound to hydroxybenzylisoproterenol and an engineered nanobody | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 4-[(1R)-1-hydroxy-2-{[1-(4-hydroxyphenyl)-2-methylpropan-2-yl]amino}ethyl]benzene-1,2-diol, Camelid Antibody Fragment, ... | | Authors: | Ring, A.M, Manglik, A, Kruse, A.C, Enos, M.D, Weis, W.I, Garcia, K.C, Kobilka, B.K. | | Deposit date: | 2013-06-24 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Adrenaline-activated structure of beta 2-adrenoceptor stabilized by an engineered nanobody.
Nature, 502, 2013
|
|
7L1J
 
 | |
8IGB
 
 | |
8IGA
 
 | |
5EIC
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with AYC | | Descriptor: | 1,2-ETHANEDIOL, 2-[(chloroacetyl)amino]-5-[(E)-(4-sulfophenyl)diazenyl]benzenesulfonic acid, CREB-binding protein | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2015-10-29 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding Motifs in the CBP Bromodomain: An Analysis of 20 Crystal Structures of Complexes with Small Molecules.
ACS Med Chem Lett, 9, 2018
|
|
5JN7
 
 | | Carbonic Anhydrase II In Complex WITH U-CH3 | | Descriptor: | 4-{[(3,5-dimethylphenyl)carbamoyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | McKenna, R, Mboge, M.Y, Mahon, B.P. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.517 Å) | | Cite: | Carbonic Anhydrase II In Complex WITH U-CH3
To Be Published
|
|
5JBH
 
 | | Cryo-EM structure of a full archaeal ribosomal translation initiation complex in the P-IN conformation | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein SX, 30S ribosomal protein eL41, ... | | Authors: | Coureux, P.-D, Schmitt, E, Mechulam, Y. | | Deposit date: | 2016-04-13 | | Release date: | 2016-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (5.34 Å) | | Cite: | Cryo-EM study of start codon selection during archaeal translation initiation.
Nat Commun, 7, 2016
|
|
7JWX
 
 | | Crystal Structure of Trypsin Bound O-methyl Benzamidine | | Descriptor: | 4-[(1-{(1S,2S)-1-[1-(4-aminobutyl)-1H-1,2,3-triazol-4-yl]-2-methylbutyl}-1H-1,2,3-triazol-4-yl)methoxy]-3-methoxybenzene-1-carboximidamide, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Packianathan, C, Laganowsky, A. | | Deposit date: | 2020-08-26 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Small molecule peptidomimetic trypsin inhibitors: validation of an EKO binding mode, but with a twist.
Org.Biomol.Chem., 20, 2022
|
|
8W47
 
 | | X-ray crystal structure of V30M-TTR in complex with isorhapontigenin | | Descriptor: | 5-[(~{E})-2-(3-methoxy-4-oxidanyl-phenyl)ethenyl]benzene-1,3-diol, SODIUM ION, Transthyretin | | Authors: | Yokoyama, T. | | Deposit date: | 2023-08-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Resveratrol Derivatives Inhibit Transthyretin Fibrillization: Structural Insights into the Interactions between Resveratrol Derivatives and Transthyretin.
J.Med.Chem., 66, 2023
|
|
7G9R
 
 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z385450668 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-(difluoromethoxy)benzene-1-sulfonamide, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-03 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
6TE6
 
 | | Crystal structure of Dot1L in complex with an inhibitor (compound 3). | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, ~{N}1-[(~{S})-(3-chlorophenyl)-pyridin-2-yl-methyl]-4-methylsulfonyl-~{N}2-pyrimidin-2-yl-benzene-1,2-diamine | | Authors: | Scheufler, C, Stauffer, F, Be, C, Moebitz, H. | | Deposit date: | 2019-11-11 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | New Potent DOT1L Inhibitors forin VivoEvaluation in Mouse.
Acs Med.Chem.Lett., 10, 2019
|
|
1XZC
 
 | |
8J1F
 
 | | GSK101 bound state of mTRPV4 | | Descriptor: | N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, Transient receptor potential cation channel subfamily V member 4 | | Authors: | Zhen, W.X, Yang, F. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
8J1B
 
 | | GSK101 and Ruthenium Red bound state of mTRPV4 | | Descriptor: | N-[(2S)-1-{4-[N-(2,4-dichlorobenzene-1-sulfonyl)-L-seryl]piperazin-1-yl}-4-methyl-1-oxopentan-2-yl]-1-benzothiophene-2-carboxamide, Transient receptor potential cation channel subfamily V member 4, ruthenium(6+) azanide pentaamino(oxido)ruthenium (1/4/2) | | Authors: | Zhen, W.X, Yang, F. | | Deposit date: | 2023-04-12 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
966C
 
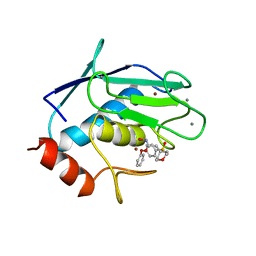 | | CRYSTAL STRUCTURE OF FIBROBLAST COLLAGENASE-1 COMPLEXED TO A DIPHENYL-ETHER SULPHONE BASED HYDROXAMIC ACID | | Descriptor: | CALCIUM ION, MMP-1, N-HYDROXY-2-[4-(4-PHENOXY-BENZENESULFONYL)-TETRAHYDRO-PYRAN-4-YL]-ACETAMIDE, ... | | Authors: | Lovejoy, B, Welch, A, Carr, S, Luong, C, Broka, C, Hendricks, R.T, Campbell, J, Walker, K, Martin, R, Van Wart, H, Browner, M.F. | | Deposit date: | 1998-08-07 | | Release date: | 1999-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors.
Nat.Struct.Biol., 6, 1999
|
|
4MQP
 
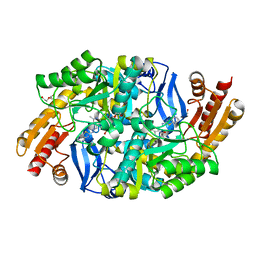 | | Mycobaterium tuberculosis transaminase BioA complexed with 2-hydrazinylbenzo[d]thiazole | | Descriptor: | (4-{[(E)-1,3-benzothiazol-2-yldiazenyl]methyl}-5-hydroxy-6-methylpyridin-3-yl)methyl dihydrogen phosphate, 1,2-ETHANEDIOL, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, ... | | Authors: | Finzel, B.C, Dai, R. | | Deposit date: | 2013-09-16 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Inhibition of Mycobacterium tuberculosis Transaminase BioA by Aryl Hydrazines and Hydrazides.
Chembiochem, 15, 2014
|
|
7KNW
 
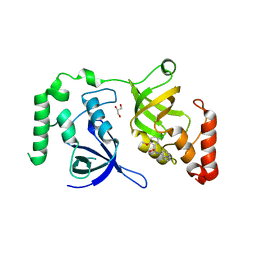 | | Crystal structure of SND1 in complex with C-26-A2 | | Descriptor: | 5-chloro-2-methoxy-N-([1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1 | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
7KNX
 
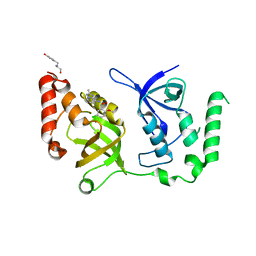 | | Crystal structure of SND1 in complex with C-26-A6 | | Descriptor: | 5-chloro-2-methoxy-N-(2-methyl[1,2,4]triazolo[1,5-a]pyridin-8-yl)benzene-1-sulfonamide, GLYCEROL, Staphylococcal nuclease domain-containing protein 1, ... | | Authors: | Kang, Y. | | Deposit date: | 2020-11-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Small-molecule inhibitors that disrupt the MTDH-SND1 complex suppress breast cancer progression and metastasis.
Nat Cancer, 3, 2022
|
|
6SPN
 
 | | Structure of the Escherichia coli methionyl-tRNA synthetase complexed with beta-methionine | | Descriptor: | (3R)-3-amino-5-(methylsulfanyl)pentanoic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Nigro, G, Schmitt, E, Mechulam, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Use of beta3-methionine as an amino acid substrate of Escherichia coli methionyl-tRNA synthetase.
J.Struct.Biol., 209, 2020
|
|
7L1P
 
 | | HIV Integrase Core domain (IN) in complex with dimer-spanning ligand | | Descriptor: | (2-{[3-(4-{2-[(3-{[3-(carboxymethyl)-5-methyl-1-benzofuran-2-yl]ethynyl}benzene-1-carbonyl)amino]ethyl}piperazine-1-carbonyl)phenyl]ethynyl}-5-methyl-1-benzofuran-3-yl)acetic acid, IODIDE ION, Integrase, ... | | Authors: | Gorman, M.A, Parker, M.W. | | Deposit date: | 2020-12-15 | | Release date: | 2021-12-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | HIV Integrase core domain in complex with inhibitor
To Be Published
|
|
7KXI
 
 | | Structure of XL5-ligated hRpn13 Pru domain | | Descriptor: | 2-{[(2S)-2-cyano-3-{3-[(4-methylbenzene-1-carbonyl)amino]phenyl}propanoyl]amino}benzoic acid, Proteasomal ubiquitin receptor ADRM1 | | Authors: | Lu, X, Walters, K.J. | | Deposit date: | 2020-12-03 | | Release date: | 2021-12-15 | | Last modified: | 2021-12-29 | | Method: | SOLUTION NMR | | Cite: | Structure-guided bifunctional molecules hit a DEUBAD-lacking hRpn13 species upregulated in multiple myeloma.
Nat Commun, 12, 2021
|
|
6SWC
 
 | | IC2B model of cryo-EM structure of a full archaeal ribosomal translation initiation complex devoid of aIF1 in P. abyssi | | Descriptor: | 16S ribosomal rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Coureux, P.-D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation.
Commun Biol, 3, 2020
|
|
