2LEW
 
 | | Structural Plasticity of Paneth cell alpha-Defensins: Characterization of Salt-Bridge Deficient Analogues of Mouse Cryptdin-4 | | Descriptor: | Alpha-defensin 4 | | Authors: | Rosengren, K, Andersson, H.S, Haugaard-Kedstrom, L.M, Bengtsson, E, Daly, N.L, Craik, D.J. | | Deposit date: | 2011-06-24 | | Release date: | 2012-05-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The alpha-defensin salt-bridge induces backbone stability to facilitate folding and confer proteolytic resistance.
Amino Acids, 43, 2012
|
|
2MAK
 
 | |
2MHP
 
 | | Solution structure of the major factor VIII binding region on von Willebrand factor | | Descriptor: | von Willebrand factor | | Authors: | Shiltagh, N, Kirkpatrick, J, Cabrita, L.D, McKinnon, T.A.J, Thalassinos, K, Tuddenham, E.G.D, Hansen, D.F. | | Deposit date: | 2013-12-02 | | Release date: | 2014-05-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major factor VIII binding region on von Willebrand factor.
Blood, 123, 2014
|
|
2LEY
 
 | | Solution structure of (R7G)-Crp4 | | Descriptor: | Alpha-defensin 4 | | Authors: | Rosengren, K, Andersson, H.S, Haugaard-Kedstrom, L.M, Bengtsson, E, Daly, N.L, Figueredo, S.M, Qu, X, Craik, D.J, Ouellette, A.J. | | Deposit date: | 2011-06-26 | | Release date: | 2012-05-16 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The alpha-defensin salt-bridge induces backbone stability to facilitate folding and confer proteolytic resistance.
Amino Acids, 43, 2012
|
|
3CP7
 
 | |
5T56
 
 | |
3E90
 
 | |
5T87
 
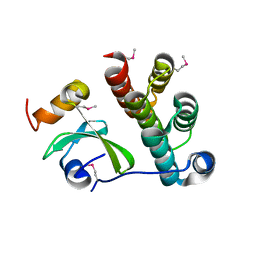 | | Crystal structure of CDI complex from Cupriavidus taiwanensis LMG 19424 | | Descriptor: | CdiA toxin, CdiI immunity protein | | Authors: | Michalska, K, Joachimiak, G, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Target highlights from the first post-PSI CASP experiment (CASP12, May-August 2016).
Proteins, 86 Suppl 1, 2018
|
|
6U2J
 
 | | EM structure of MPEG-1 (L425K, alpha conformation) soluble pre-pore complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Pang, S.S, Bayly-Jones, C. | | Deposit date: | 2019-08-20 | | Release date: | 2019-09-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1.
Nat Commun, 10, 2019
|
|
6U7S
 
 | |
6U2W
 
 | | EM structure of MPEG-1(L425K) pre-pore complex bound to liposome | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Pang, S.S, Bayly-Jones, C. | | Deposit date: | 2019-08-20 | | Release date: | 2019-09-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1.
Nat Commun, 10, 2019
|
|
6U7W
 
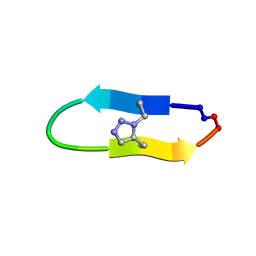 | | NMR solution structure of a triazole bridged KLK7 inhibitor | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-LYS-ALA-LEU-PHE-SER-ASN-PRO-PRO-ILE-ALA-PHE-PRO-ASN | | Authors: | White, A.M, Harvey, P.J, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2020-07-15 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6UFN
 
 | |
6UHV
 
 | |
6UII
 
 | |
6U23
 
 | | EM structure of MPEG-1(w.t.) soluble pre-pore | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Pang, S.S, Bayly-Jones, C. | | Deposit date: | 2019-08-19 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1.
Nat Commun, 10, 2019
|
|
6U2L
 
 | | EM structure of MPEG-1 (L425K, beta conformation) soluble pre-pore complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Pang, S.S, Bayly-Jones, C. | | Deposit date: | 2019-08-20 | | Release date: | 2019-09-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1.
Nat Commun, 10, 2019
|
|
6U2K
 
 | |
6UIM
 
 | |
6UIJ
 
 | |
6U7X
 
 | | NMR solution structure of triazole bridged plasmin inhibitor | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-ARG-ALA-TYR-LYS-SER-LYS-PRO-PRO-ILE-ALA-PHE-PRO-ASP | | Authors: | White, A.M, Harvey, P.J, Wang, C.K, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6UHU
 
 | |
5NLF
 
 | | Crystal structure of Zn2.7-E16V human ubiquitin (hUb) mutant adduct, from a solution 100 mM zinc acetate/1.3 mM E16V hUb | | Descriptor: | ACETATE ION, Polyubiquitin-C, ZINC ION | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
3LJ6
 
 | |
3LJ7
 
 | |
