4AP6
 
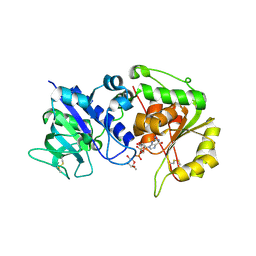 | | Crystal structure of human POFUT2 E54A mutant in complex with GDP- fucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GDP-FUCOSE PROTEIN O-FUCOSYLTRANSFERASE 2, ... | | Authors: | Chen, C, Keusch, J.J, Klein, D, Hess, D, Hofsteenge, J, Gut, H. | | Deposit date: | 2012-03-30 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Structure of Human Pofut2: Insights Into Thrombospondin Type 1 Repeat Fold and O-Fucosylation.
Embo J., 31, 2012
|
|
1M4X
 
 | | PBCV-1 virus capsid, quasi-atomic model | | Descriptor: | PBCV-1 virus capsid | | Authors: | Nandhagopal, N, Simpson, A.A, Gurnon, J.R, Yan, X, Baker, T.S, Graves, M.V, Van Etten, J.L, Rossmann, M.G. | | Deposit date: | 2002-07-05 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | The Structure and Evolution of the Major Capsid Protein of a Large,
Lipid containing, DNA virus.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3RC2
 
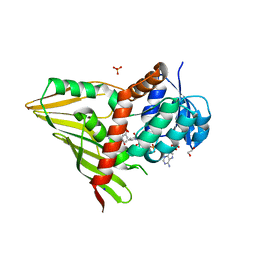 | | Crystal Structure of KijD10, a 3-ketoreductase from Actinomadura kijaniata in complex with TDP-benzene and NADP; open conformation | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phenoxy)phosphoryl]oxy}phosphoryl]thymidine, CHLORIDE ION, ... | | Authors: | Holden, H.M, Kubiak, R.L. | | Deposit date: | 2011-03-30 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Combined Structural and Functional Investigation of a C-3''-Ketoreductase Involved in the Biosynthesis of dTDP-l-Digitoxose.
Biochemistry, 50, 2011
|
|
2O93
 
 | |
1GDV
 
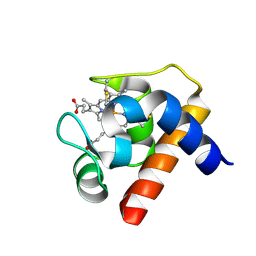 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 FROM RED ALGA PORPHYRA YEZOENSIS AT 1.57 A RESOLUTION | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Yamada, S, Park, S.-Y, Shimizu, H, Shiro, Y, Oku, T. | | Deposit date: | 2000-10-06 | | Release date: | 2001-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure of cytochrome c6 from the red alga Porphyra yezoensis at 1. 57 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
4MHY
 
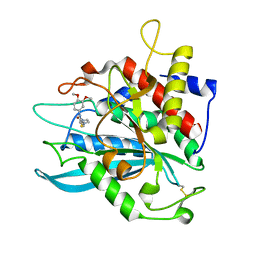 | | Crystal structure of a glutaminyl cyclase from Ixodes scapularis in complex with PBD150 | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, Glutaminyl cyclase, putative, ... | | Authors: | Huang, K.F, Hsu, H.L, Wang, A.H.J. | | Deposit date: | 2013-08-30 | | Release date: | 2014-03-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural and functional analyses of a glutaminyl cyclase from Ixodes scapularis reveal metal-independent catalysis and inhibitor binding.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1FW4
 
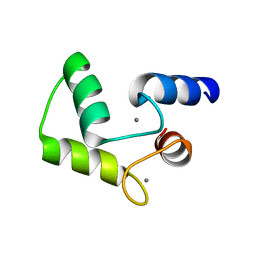 | |
3S9J
 
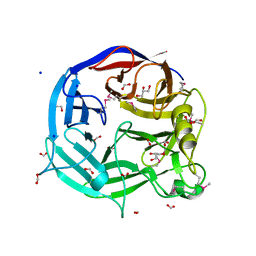 | |
3VZY
 
 | | Crystal structure of PcrB complexed with G1P from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
2O6C
 
 | | Structure of selenomethionyl rTp34 from Treponema pallidum | | Descriptor: | 1,2-ETHANEDIOL, 34 kDa membrane antigen, CHLORIDE ION, ... | | Authors: | Machius, M, Brautigam, C.A, Deka, R.K, Tomchick, D.R, Lumpkins, S.B, Norgard, M.V. | | Deposit date: | 2006-12-07 | | Release date: | 2006-12-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the Tp34 (TP0971) lipoprotein of treponema pallidum: implications of its metal-bound state and affinity for human lactoferrin.
J.Biol.Chem., 282, 2007
|
|
2C5A
 
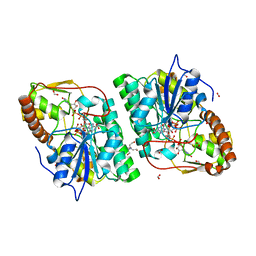 | | GDP-mannose-3', 5' -epimerase (Arabidopsis thaliana),Y174F, with GDP-beta-L-galactose bound in the active site | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, GDP-MANNOSE-3', ... | | Authors: | Major, L.L, Wolucka, B.A, Naismith, J.H. | | Deposit date: | 2005-10-26 | | Release date: | 2005-11-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and Function of Gdp-Mannose-3',5'-Epimerase: An Enzyme which Performs Three Chemical Reactions at the Same Active Site.
J.Am.Chem.Soc., 127, 2005
|
|
2YVL
 
 | |
3SLC
 
 | |
1G5C
 
 | | CRYSTAL STRUCTURE OF THE 'CAB' TYPE BETA CLASS CARBONIC ANHYDRASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-CARBONIC ANHYDRASE, CALCIUM ION, ... | | Authors: | Strop, P, Smith, K.S, Iverson, T.M, Ferry, J.G, Rees, D.C. | | Deposit date: | 2000-10-31 | | Release date: | 2001-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the "cab"-type beta class carbonic anhydrase from the archaeon Methanobacterium thermoautotrophicum.
J.Biol.Chem., 276, 2001
|
|
3PA0
 
 | | Crystal Structure of Chiral Gamma-PNA with Complementary DNA Strand: Insight Into the Stability and Specificity of Recognition an Conformational Preorganization | | Descriptor: | 1,2-ETHANEDIOL, DNA 5'-D(*AP*TP*CP*TP*GP*TP*GP*GP*TP*C)-3', PEPTIDE NUCLEIC ACID, ... | | Authors: | Yeh, J.I, Shivachev, B, Du, S. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of chiral gammaPNA with complementary DNA strand: insights into the stability and specificity of recognition and conformational preorganization.
J.Am.Chem.Soc., 132, 2010
|
|
2A0Y
 
 | | Structure of human purine nucleoside phosphorylase H257D mutant | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Murkin, A.S, Shi, W, Schramm, V.L. | | Deposit date: | 2005-06-17 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Neighboring group participation in the transition state of human purine nucleoside phosphorylase.
Biochemistry, 46, 2007
|
|
3H0W
 
 | | Human AdoMetDC with 5'-Deoxy-5'-[(N-dimethyl)amino]-8-methyl-adenosine | | Descriptor: | 1,4-DIAMINOBUTANE, 5'-deoxy-5'-(dimethylamino)-8-methyladenosine, PYRUVIC ACID, ... | | Authors: | Bale, S, Brooks, W.H, Hanes, J.W, Mahesan, A.M, Guida, W.C, Ealick, S.E. | | Deposit date: | 2009-04-10 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Role of the sulfonium center in determining the ligand specificity of human s-adenosylmethionine decarboxylase.
Biochemistry, 48, 2009
|
|
3W10
 
 | |
7LXI
 
 | |
2P27
 
 | | Crystal Structure of Human Pyridoxal Phosphate Phosphatase with Mg2+ at 1.9 A resolution | | Descriptor: | MAGNESIUM ION, Pyridoxal phosphate phosphatase | | Authors: | Ramagopal, U.A, Freeman, J, Izuka, M, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-07 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
4INX
 
 | | Structure of Pheromone-binding protein 1 in complex with (Z,Z)-11,13- hexadecadienol | | Descriptor: | (11Z,13Z)-hexadeca-11,13-dien-1-ol, Pheromone-binding protein 1 | | Authors: | di Luccio, E, Wilson, D.K. | | Deposit date: | 2013-01-07 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.853 Å) | | Cite: | Crystallographic Observation of pH-Induced Conformational Changes in the Amyelois transitella Pheromone-Binding Protein AtraPBP1.
Plos One, 8, 2013
|
|
4QT1
 
 | |
5STD
 
 | | SCYTALONE DEHYDRATASE PLUS INHIBITOR 2 | | Descriptor: | (6,7-DIFLUORO-QUINAZOLIN-4-YL)-(1-METHYL-2,2-DIPHENYL-ETHYL)-AMINE, CALCIUM ION, Scytalone dehydratase | | Authors: | Wawrzak, Z, Sandalova, T, Steffens, J.J, Basarab, G.S, Lundqvist, T, Lindqvist, Y, Jordan, D.B. | | Deposit date: | 1999-02-10 | | Release date: | 1999-12-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution structures of scytalone dehydratase-inhibitor complexes crystallized at physiological pH.
Proteins, 35, 1999
|
|
1YAJ
 
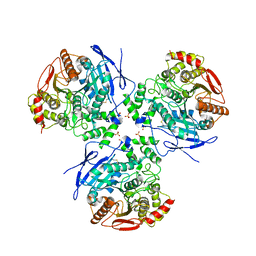 | | Crystal Structure of Human Liver Carboxylesterase in complex with benzil | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BENZOIC ACID, CES1 protein, ... | | Authors: | Fleming, C.D, Bencharit, S, Edwards, C.C, Hyatt, J.L, Morton, C.M, Howard-Williams, E.L, Potter, P.M, Redinbo, M.R. | | Deposit date: | 2004-12-17 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into drug processing by human carboxylesterase 1: tamoxifen, mevastatin, and inhibition by benzil.
J.Mol.Biol., 352, 2005
|
|
2PA4
 
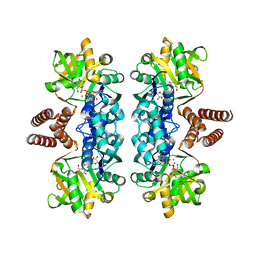 | |
