8A9F
 
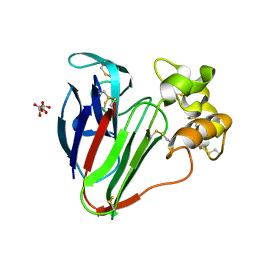 | |
6VXX
 
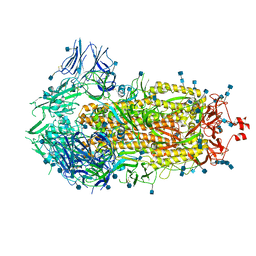 | | Structure of the SARS-CoV-2 spike glycoprotein (closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Walls, A.C, Park, Y.J, Tortorici, M.A, Wall, A, Seattle Structural Genomics Center for Infectious Disease (SSGCID), McGuire, A.T, Veesler, D. | | Deposit date: | 2020-02-25 | | Release date: | 2020-03-11 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein.
Cell, 181, 2020
|
|
6W2T
 
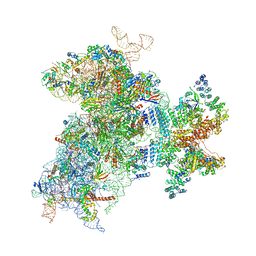 | | Structure of the Cricket Paralysis Virus 5-UTR IRES (CrPV 5-UTR-IRES) bound to the small ribosomal subunit in the closed state (Class 2) | | Descriptor: | 18S rRNA, CrPV 5'-UTR IRES, Eukaryotic translation initiation factor 3 subunit A, ... | | Authors: | Neupane, R, Pisareva, V, Rodriguez, C.F, Pisarev, A, Fernandez, I.S. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | A complex IRES at the 5'-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate.
Elife, 9, 2020
|
|
8AEN
 
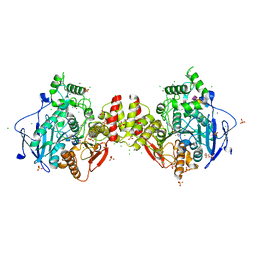 | | Human acetylcholinesterase in complex with zinc and N,N,N-trimethyl-2-oxo-2-(2-(pyridin-2-ylmethylene)hydrazineyl)ethan-1-aminium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Nachon, F, Dias, J, Brazzolotto, X. | | Deposit date: | 2022-07-13 | | Release date: | 2023-06-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Grid-Type Quaternary Metallosupramolecular Compounds Inhibit Human Cholinesterases through Dynamic Multivalent Interactions.
Chembiochem, 23, 2022
|
|
7THK
 
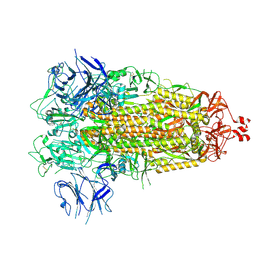 | |
7TGH
 
 | | Cryo-EM structure of respiratory super-complex CI+III2 from Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | Authors: | Zhou, L, Maldonado, M, Padavannil, A, Guo, F, Letts, J.A. | | Deposit date: | 2022-01-07 | | Release date: | 2022-04-06 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of Tetrahymena 's respiratory chain reveal the diversity of eukaryotic core metabolism.
Science, 376, 2022
|
|
7U97
 
 | | SAAV pH 4.0 capsid structure | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
6VZR
 
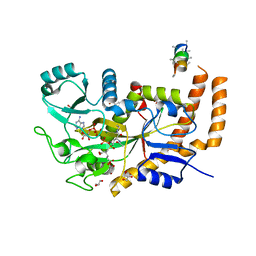 | | Engineered TTLL6 bound to the initiation analog | | Descriptor: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-(ethylamino)-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8APO
 
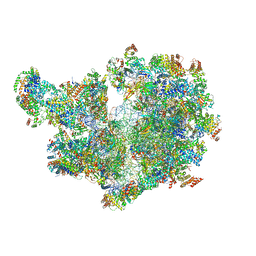 | | Structure of the mitochondrial ribosome from Polytomella magna with tRNAs bound to the A and P sites | | Descriptor: | A-site tRNA anticodon loop, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tobiasson, V, Berzina, I, Amunts, A. | | Deposit date: | 2022-08-10 | | Release date: | 2023-06-21 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a mitochondrial ribosome with fragmented rRNA in complex with membrane-targeting elements.
Nat Commun, 13, 2022
|
|
7U95
 
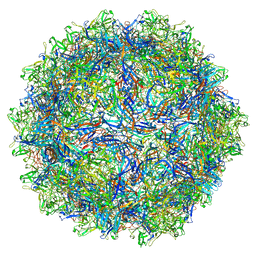 | | SAAV pH 6.0 capsid structure | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
6W0F
 
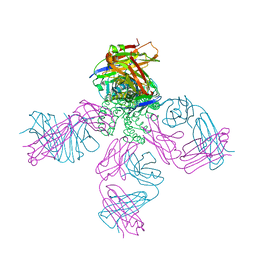 | | Closed-gate KcsA soaked in 0mM KCl/5mM BaCl2 | | Descriptor: | Fab Heavy Chain, Fab Light Chain, POTASSIUM ION, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
7TJA
 
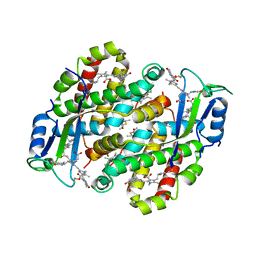 | | Structure of the Light Harvesting Complex PE545 from Proteomonas sulcata | | Descriptor: | 15,16-DIHYDROBILIVERDIN, MAGNESIUM ION, PHYCOERYTHROBILIN, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2022-01-15 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
6W9G
 
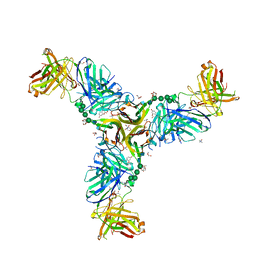 | | Crystal Structure of the Fab fragment of humanized 5c8 antibody containing the fluorescent non-canonical amino acid L-(7-hydroxycoumarin-4-yl)ethylglycine in complex with CD40L at pH 6.8 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 5c8* Fab (heavy chain), ... | | Authors: | Henderson, J.N, Simmons, C.R, Mills, J.H. | | Deposit date: | 2020-03-23 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Insights into How Protein Environments Tune the Spectroscopic Properties of a Noncanonical Amino Acid Fluorophore.
Biochemistry, 59, 2020
|
|
6W1U
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7U96
 
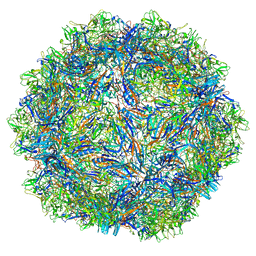 | | SAAV pH 5.5 capsid structure | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
7U94
 
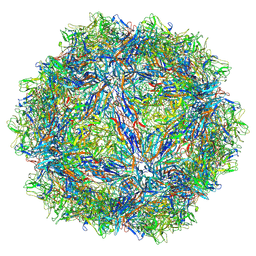 | | SAAV pH 7.4 capsid structure | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2022-03-10 | | Release date: | 2022-04-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Characterization of the Serpentine Adeno-Associated Virus (SAAV) Capsid Structure: Receptor Interactions and Antigenicity.
J.Virol., 96, 2022
|
|
6W4E
 
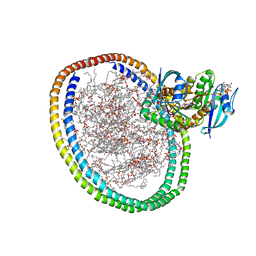 | | NMR-driven structure of KRAS4B-GTP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6WBG
 
 | | Cryo-EM structure of human Pannexin 1 channel with its C-terminal tail cleaved by caspase-7 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Lu, W, Du, J, Ruan, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-03 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of human pannexin 1 reveal ion pathways and mechanism of gating.
Nature, 584, 2020
|
|
6WBF
 
 | | Cryo-EM structure of wild type human Pannexin 1 channel | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Lu, W, Du, J, Ruan, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-03 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structures of human pannexin 1 reveal ion pathways and mechanism of gating.
Nature, 584, 2020
|
|
6VR0
 
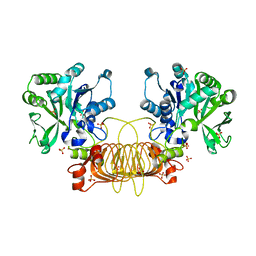 | | Agrobacterium Tumefaciens ADP-glucose pyrophosphorylase W106A | | Descriptor: | GLYCEROL, Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Mascarenhas, R.N, Liu, D, Ballicora, M, Iglesias, A, Asencion, M, Figueroa, C. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Agrobacterium Tumefaciens ADP-glucose pyrophosphorylase W106A
To Be Published
|
|
8ARI
 
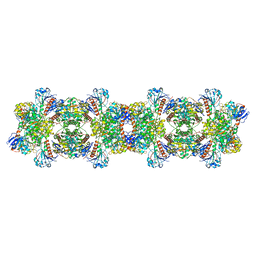 | |
7TQL
 
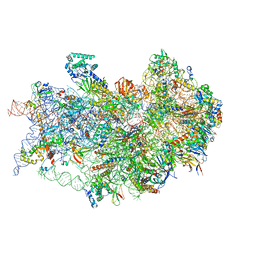 | | CryoEM structure of the human 40S small ribosomal subunit in complex with translation initiation factors eIF1A and eIF5B. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Lapointe, C.P, Grosely, R, Sokabe, M, Alvarado, C, Wang, J, Montabana, E, Villa, N, Shin, B, Dever, T, Fraser, C, Fernandez, I.S, Puglisi, J.D. | | Deposit date: | 2022-01-26 | | Release date: | 2022-04-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | eIF5B and eIF1A reorient initiator tRNA to allow ribosomal subunit joining.
Nature, 607, 2022
|
|
8B2L
 
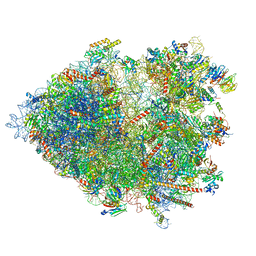 | | Cryo-EM structure of the plant 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 30S ribosomal protein S15, ... | | Authors: | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | Deposit date: | 2022-09-14 | | Release date: | 2023-08-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
7TMZ
 
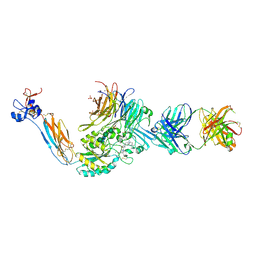 | | Integrin alaphIIBbeta3 complex with BMS compound 4 | | Descriptor: | (4-{[(5S)-3-{4-[(E)-imino(4-methylpiperazin-1-yl)methyl]phenyl}-4,5-dihydro-1,2-oxazol-5-yl]methyl}piperazin-1-yl)acetic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhu, J, Lin, F.-Y, Zhu, J, Springer, T.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.20002 Å) | | Cite: | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
8ASA
 
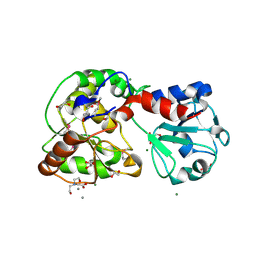 | | Crystal structure of AO75L | | Descriptor: | 1,2-ETHANEDIOL, BICINE, CALCIUM ION, ... | | Authors: | Laugeri, M.E, Speciale, I, Gimeno, A, Lin, S, Poveda, A, Lowary, T, Van Etten, J.L, Barbero, J.J, De Castro, C, Tonetti, M, Rojas, A.L. | | Deposit date: | 2022-08-18 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of AO75L
To Be Published
|
|
