7S5F
 
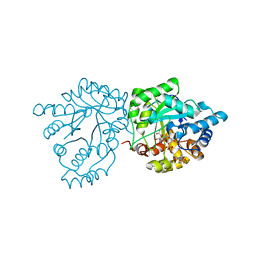 | | Crystal structure of mannose-6-phosphate reductase from celery (Apium graveolens) leaves with NADP+ and mannonic acid bound | | Descriptor: | D-MANNONIC ACID, Manose-6-phosphate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, Y, Bhayani, J.A, Romina, I.M, Hartman, M.D, Cereijo, A.E, Ballicora, M.A, Iglesias, A.A, Figueroa, C.M, Liu, D. | | Deposit date: | 2021-09-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Determinants of Sugar Alcohol Biosynthesis in Plants: The Crystal Structures of Mannose-6-Phosphate and Aldose-6-Phosphate Reductases.
Plant Cell.Physiol., 63, 2022
|
|
7S5I
 
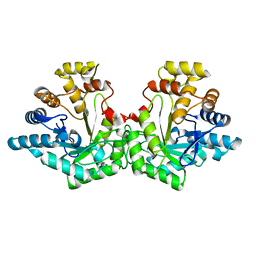 | | Crystal structure of Aldose-6-phosphate reductase (Ald6PRase) from peach (Prunus persica) leaves | | Descriptor: | Sorbitol-6-phosphate dehydrogenase | | Authors: | Zheng, Y, Bhayani, J.A, Romina, I.M, Hartman, M.D, Cereijo, A.E, Ballicora, M.A, Iglesias, A.A, Figueroa, C.M, Liu, D. | | Deposit date: | 2021-09-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural Determinants of Sugar Alcohol Biosynthesis in Plants: The Crystal Structures of Mannose-6-Phosphate and Aldose-6-Phosphate Reductases.
Plant Cell.Physiol., 63, 2022
|
|
6M8H
 
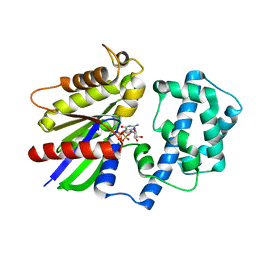 | | Crystal Structure of the R208Q mutant of G(i) subunit alpha-1 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, MAGNESIUM ION | | Authors: | Mascarenhas, R, Goossens, J, Leverson, B, Kothawala, S, Ballicora, M, Olsen, K, de freitas, D, Liu, D. | | Deposit date: | 2018-08-21 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | FUNCTIONAL CONSEQUENCES OF ONCOGENIC MUTATIONS IN THE SWITCH II REGION OF Galphai1 and Galphas PROTEINS
To Be Published
|
|
4RBN
 
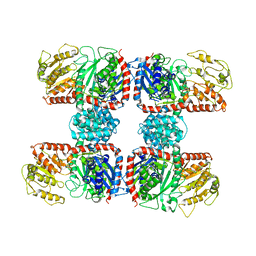 | | The crystal structure of Nitrosomonas europaea sucrose synthase: Insights into the evolutionary origin of sucrose metabolism in prokaryotes | | Descriptor: | Sucrose synthase:Glycosyl transferases group 1 | | Authors: | Wu, R, Asencion Diez, M.D, Figueroa, C.M, Machtey, M, Iglesias, A.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2014-09-12 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The Crystal Structure of Nitrosomonas europaea Sucrose Synthase Reveals Critical Conformational Changes and Insights into Sucrose Metabolism in Prokaryotes.
J.Bacteriol., 197, 2015
|
|
6V96
 
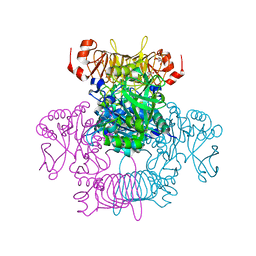 | | Agrobacterium tumefaciens ADP-Glucose pyrophosphorylase-S72E | | Descriptor: | CITRIC ACID, GLYCEROL, Glucose-1-phosphate adenylyltransferase | | Authors: | Zheng, Y, Hussien, R, Alghamdi, M.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Site-directed mutagenesis of Serine-72 reveals the location of the fructose 6-phosphate regulatory site of the Agrobacterium tumefaciens ADP-glucose pyrophosphorylase.
Protein Sci., 31, 2022
|
|
6V99
 
 | | Agrobacterium tumefaciens ADP-Glucose pyrophosphorylase- S72D in the presence of sulfate | | Descriptor: | Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Zheng, Y, Alghamdi, M.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Site-directed mutagenesis of Serine-72 reveals the location of the fructose 6-phosphate regulatory site of the Agrobacterium tumefaciens ADP-glucose pyrophosphorylase.
Protein Sci., 31, 2022
|
|
6V9A
 
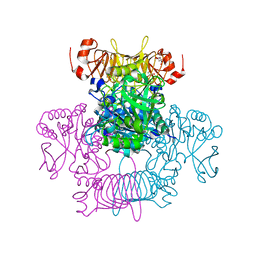 | | Agrobacterium tumefaciens ADP-Glucose pyrophosphorylase-S72D | | Descriptor: | CITRIC ACID, GLYCEROL, Glucose-1-phosphate adenylyltransferase | | Authors: | Zheng, Y, Alghamdi, M.A, Ballicora, M.A, Liu, D. | | Deposit date: | 2019-12-13 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Site-directed mutagenesis of Serine-72 reveals the location of the fructose 6-phosphate regulatory site of the Agrobacterium tumefaciens ADP-glucose pyrophosphorylase.
Protein Sci., 31, 2022
|
|
6VR0
 
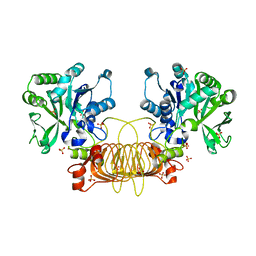 | | Agrobacterium Tumefaciens ADP-glucose pyrophosphorylase W106A | | Descriptor: | GLYCEROL, Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Mascarenhas, R.N, Liu, D, Ballicora, M, Iglesias, A, Asencion, M, Figueroa, C. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Agrobacterium Tumefaciens ADP-glucose pyrophosphorylase W106A
To Be Published
|
|
5W5T
 
 | | Agrobacterium tumefaciens ADP-Glucose Pyrophosphorylase bound to activator ethyl pyruvate | | Descriptor: | GLYCEROL, Glucose-1-phosphate adenylyltransferase, SULFATE ION, ... | | Authors: | Mascarenhas, R.N, Hill, B.L, Ballicora, M.A, Liu, D. | | Deposit date: | 2017-06-15 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural analysis reveals a pyruvate-binding activator site in theAgrobacterium tumefaciensADP-glucose pyrophosphorylase.
J. Biol. Chem., 294, 2019
|
|
5W6J
 
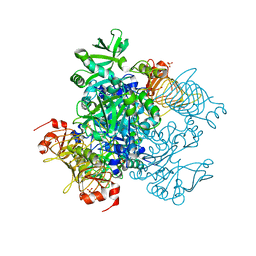 | | Agrobacterium tumefaciens ADP-glucose pyrophosphorylase | | Descriptor: | Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Mascarenhas, R.N, Hill, B.L, Wu, R, Ballicora, M.A, Liu, D. | | Deposit date: | 2017-06-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural analysis reveals a pyruvate-binding activator site in theAgrobacterium tumefaciensADP-glucose pyrophosphorylase.
J. Biol. Chem., 294, 2019
|
|
5W5R
 
 | | Agrobacterium tumefaciens ADP-glucose pyrophosphorylase P96A mutant bound to activator pyruvate | | Descriptor: | Glucose-1-phosphate adenylyltransferase, PYRUVIC ACID, SULFATE ION | | Authors: | Mascarenhas, R.N, Hill, B.L, Ballicora, M.A, Liu, D. | | Deposit date: | 2017-06-15 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Structural analysis reveals a pyruvate-binding activator site in theAgrobacterium tumefaciensADP-glucose pyrophosphorylase.
J. Biol. Chem., 294, 2019
|
|
1YP2
 
 | | Crystal structure of potato tuber ADP-glucose pyrophosphorylase | | Descriptor: | Glucose-1-phosphate adenylyltransferase small subunit, PARA-MERCURY-BENZENESULFONIC ACID, SULFATE ION | | Authors: | Jin, X, Ballicora, M.A, Preiss, J, Geiger, J.H. | | Deposit date: | 2005-01-29 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of potato tuber ADP-glucose pyrophosphorylase.
Embo J., 24, 2005
|
|
1YP3
 
 | | Crystal structure of potato tuber ADP-glucose pyrophosphorylase in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Glucose-1-phosphate adenylyltransferase small subunit, SULFATE ION | | Authors: | Jin, X, Ballicora, M.A, Preiss, J, Geiger, J.H. | | Deposit date: | 2005-01-29 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of potato tuber ADP-glucose pyrophosphorylase.
Embo J., 24, 2005
|
|
1YP4
 
 | | Crystal structure of potato tuber ADP-glucose pyrophosphorylase in complex with ADP-glucose | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, Glucose-1-phosphate adenylyltransferase small subunit, ... | | Authors: | Jin, X, Ballicora, M.A, Preiss, J, Geiger, J.H. | | Deposit date: | 2005-01-29 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of potato tuber ADP-glucose pyrophosphorylase.
Embo J., 24, 2005
|
|
