8STW
 
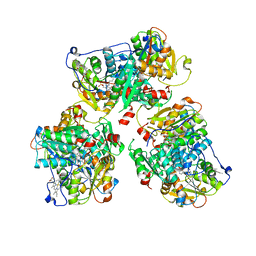 | | K384N HUMAN CYSTATHIONINE BETA-SYNTHASE (delta 411-551) | | Descriptor: | Cystathionine beta-synthase, K384N variant, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mascarenhas, R, Roman, J, Banerjee, R. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Disease-causing cystathionine beta-synthase linker mutations impair allosteric regulation.
J.Biol.Chem., 299, 2023
|
|
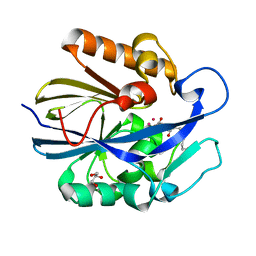
7L5F
 
 | |
6X8Z
 
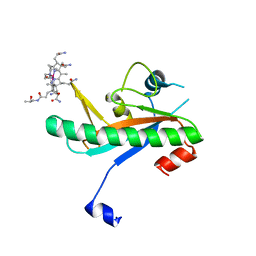 | | Crystal structure of N-truncated human B12 chaperone CblD(C262S)-thiolato-cob(III)alamin complex (108-296) | | Descriptor: | COBALAMIN, Methylmalonic aciduria and homocystinuria type D protein, mitochondrial | | Authors: | Mascarenhas, R, Li, Z, Koutmos, M, Banerjee, R. | | Deposit date: | 2020-06-02 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Interprotein Co-S Coordination Complex in the B 12 -Trafficking Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
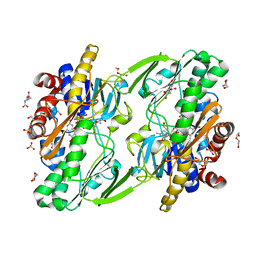
8DHK
 
 | |
8GJU
 
 | | Crystal structure of human methylmalonyl-CoA mutase (MMUT) in complex with methylmalonic acidemia type A protein (MMAA), coenzyme A, and GDP | | Descriptor: | COENZYME A, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mascarenhas, R.M, Ruetz, M, Gouda, H, Yaw, M, Banerjee, R. | | Deposit date: | 2023-03-16 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Architecture of the human G-protein-methylmalonyl-CoA mutase nanoassembly for B 12 delivery and repair.
Nat Commun, 14, 2023
|
|
8DYL
 
 | | Crystal structure of human methylmalonyl-CoA mutase bound to aquocobalamin | | Descriptor: | COBALAMIN, GLYCEROL, Methylmalonyl-CoA mutase, ... | | Authors: | Mascarenhas, R.N, Gouda, H, Banerjee, R. | | Deposit date: | 2022-08-04 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bivalent molecular mimicry by ADP protects metal redox state and promotes coenzyme B 12 repair.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DYJ
 
 | | Crystal structure of human methylmalonyl-CoA mutase in complex with ADP and cob(II)alamin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COBALAMIN, Methylmalonyl-CoA mutase, ... | | Authors: | Mascarenhas, R.N, Gouda, H, Banerjee, R. | | Deposit date: | 2022-08-04 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bivalent molecular mimicry by ADP protects metal redox state and promotes coenzyme B 12 repair.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8D32
 
 | | Mycobacterium tuberculosis pduO-type ATP:cobalamin adenosyltransferase bound to 5-deoxyadenosylrhodibalamin and PPPi | | Descriptor: | 5'-DEOXYADENOSINE, Corrinoid adenosyltransferase, MAGNESIUM ION, ... | | Authors: | Mascarenhas, R.N, Ruetz, M, Koutmos, M, Banerjee, R. | | Deposit date: | 2022-05-31 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A noble substitution leads to the cofactor mimicry by rhodibalamin
To Be Published
|
|
7RUU
 
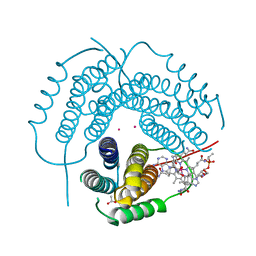 | | Structure of Human ATP:Cobalamin Adenosyltransferase R190C bound to adenosylcobalamin | | Descriptor: | 5'-DEOXYADENOSINE, ACETATE ION, COBALAMIN, ... | | Authors: | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | Deposit date: | 2021-08-18 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
7RUT
 
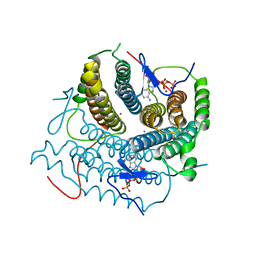 | | Structure of Human ATP:Cobalamin Adenosyltransferase R190C bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Corrinoid adenosyltransferase, GLYCEROL, ... | | Authors: | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | Deposit date: | 2021-08-18 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
7RUV
 
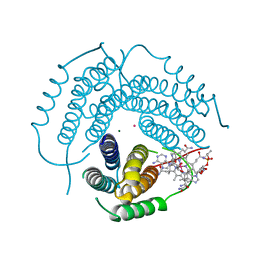 | | Structure of Human ATP:Cobalamin Adenosyltransferase E193K bound to adenosylcobalamin | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Corrinoid adenosyltransferase, ... | | Authors: | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | Deposit date: | 2021-08-18 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
6WH5
 
 | | Mycobacterium tuberculosis pduO-type ATP:cobalamin adenosyltransferase bound to cob(II)alamin and PPPi | | Descriptor: | COBALAMIN, Corrinoid adenosyltransferase, MAGNESIUM ION, ... | | Authors: | Mascarenhas, R.N, Ruetz, M, Koutmos, M, Banerjee, R. | | Deposit date: | 2020-04-07 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | Mobile loop dynamics in adenosyltransferase control binding and reactivity of coenzyme B 12 .
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WGV
 
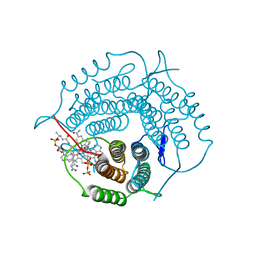 | | Mycobacterium tuberculosis pduO-type ATP:cobalamin adenosyltransferase bound to adenosylcobalamin and PPPi | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Corrinoid adenosyltransferase, ... | | Authors: | Mascarenhas, R.N, Ruetz, M, Koutmos, M, Banerjee, R. | | Deposit date: | 2020-04-06 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Mobile loop dynamics in adenosyltransferase control binding and reactivity of coenzyme B 12 .
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WGS
 
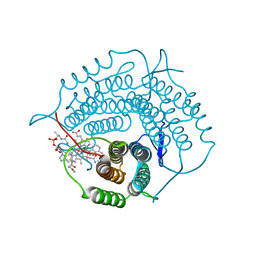 | | Mycobacterium tuberculosis pduO-type ATP:cobalamin adenosyltransferase bound to adenosylcobalamin | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Corrinoid adenosyltransferase | | Authors: | Mascarenhas, R.N, Ruetz, M, Koutmos, M, Banerjee, R. | | Deposit date: | 2020-04-06 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mobile loop dynamics in adenosyltransferase control binding and reactivity of coenzyme B 12 .
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WGU
 
 | | Mycobacterium tuberculosis pduO-type ATP:cobalamin adenosyltransferase | | Descriptor: | Corrinoid adenosyltransferase, SULFATE ION | | Authors: | Mascarenhas, R.N, Ruetz, M, Koutmos, M, Banerjee, R. | | Deposit date: | 2020-04-06 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mobile loop dynamics in adenosyltransferase control binding and reactivity of coenzyme B 12 .
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4ZO2
 
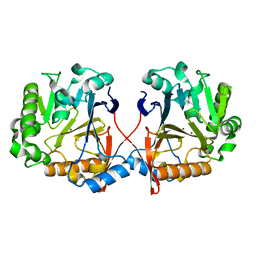 | | AidC, a Dizinc Quorum-Quenching Lactonase | | Descriptor: | Acylhomoserine lactonase, ZINC ION | | Authors: | Mascarenhas, R, Thomas, P.W, Wu, C.-X, Nocek, B.P, Hoang, Q, Fast, W, Liu, D. | | Deposit date: | 2015-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Structural and Biochemical Characterization of AidC, a Quorum-Quenching Lactonase with Atypical Selectivity.
Biochemistry, 54, 2015
|
|
4ZO3
 
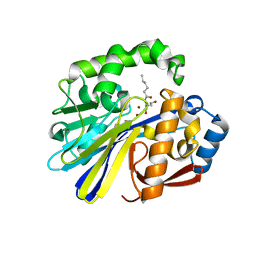 | | AidC, a Dizinc Quorum-Quenching Lactonase, in complex with a product N-hexnoyl-L-homoserine | | Descriptor: | Acylhomoserine lactonase, N-hexanoyl-L-homoserine, ZINC ION | | Authors: | Mascarenhas, R, Thomas, P.W, Wu, C.-X, Nocek, B.P, Hoang, Q, Fast, W, Liu, D. | | Deposit date: | 2015-05-05 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Biochemical Characterization of AidC, a Quorum-Quenching Lactonase with Atypical Selectivity.
Biochemistry, 54, 2015
|
|
6M8H
 
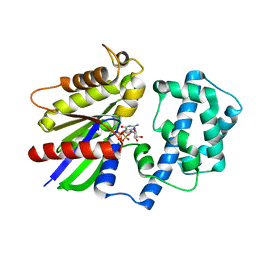 | | Crystal Structure of the R208Q mutant of G(i) subunit alpha-1 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1, MAGNESIUM ION | | Authors: | Mascarenhas, R, Goossens, J, Leverson, B, Kothawala, S, Ballicora, M, Olsen, K, de freitas, D, Liu, D. | | Deposit date: | 2018-08-21 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | FUNCTIONAL CONSEQUENCES OF ONCOGENIC MUTATIONS IN THE SWITCH II REGION OF Galphai1 and Galphas PROTEINS
To Be Published
|
|
5W5T
 
 | | Agrobacterium tumefaciens ADP-Glucose Pyrophosphorylase bound to activator ethyl pyruvate | | Descriptor: | GLYCEROL, Glucose-1-phosphate adenylyltransferase, SULFATE ION, ... | | Authors: | Mascarenhas, R.N, Hill, B.L, Ballicora, M.A, Liu, D. | | Deposit date: | 2017-06-15 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural analysis reveals a pyruvate-binding activator site in theAgrobacterium tumefaciensADP-glucose pyrophosphorylase.
J. Biol. Chem., 294, 2019
|
|
5W6J
 
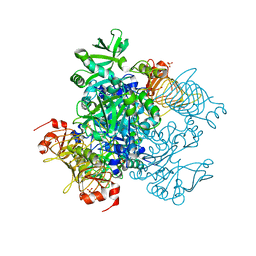 | | Agrobacterium tumefaciens ADP-glucose pyrophosphorylase | | Descriptor: | Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Mascarenhas, R.N, Hill, B.L, Wu, R, Ballicora, M.A, Liu, D. | | Deposit date: | 2017-06-16 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural analysis reveals a pyruvate-binding activator site in theAgrobacterium tumefaciensADP-glucose pyrophosphorylase.
J. Biol. Chem., 294, 2019
|
|
6VR0
 
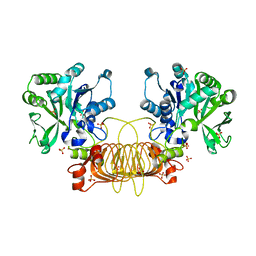 | | Agrobacterium Tumefaciens ADP-glucose pyrophosphorylase W106A | | Descriptor: | GLYCEROL, Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Mascarenhas, R.N, Liu, D, Ballicora, M, Iglesias, A, Asencion, M, Figueroa, C. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Agrobacterium Tumefaciens ADP-glucose pyrophosphorylase W106A
To Be Published
|
|
6B6G
 
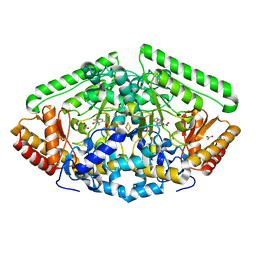 | | Crystal Structure of GABA Aminotransferase bound to (S)-3-Amino-4-(difluoromethylenyl)cyclopent-1-ene-1-carboxylic acid, an Potent Inactivatorfor the Treatment of Addiction | | Descriptor: | (3R,4E)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]cyclopent-1-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Mascarenhas, R, Juncosa, J.I, Takaya, K, Le, L.V, Moschitto, M.J, Silverman, R.B, Liu, D. | | Deposit date: | 2017-10-02 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design and Mechanism of (S)-3-Amino-4-(difluoromethylenyl)cyclopent-1-ene-1-carboxylic Acid, a Highly Potent gamma-Aminobutyric Acid Aminotransferase Inactivator for the Treatment of Addiction.
J. Am. Chem. Soc., 140, 2018
|
|
5W5R
 
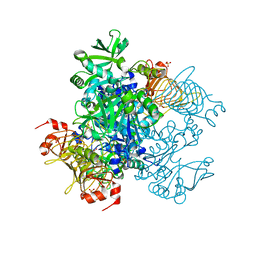 | | Agrobacterium tumefaciens ADP-glucose pyrophosphorylase P96A mutant bound to activator pyruvate | | Descriptor: | Glucose-1-phosphate adenylyltransferase, PYRUVIC ACID, SULFATE ION | | Authors: | Mascarenhas, R.N, Hill, B.L, Ballicora, M.A, Liu, D. | | Deposit date: | 2017-06-15 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Structural analysis reveals a pyruvate-binding activator site in theAgrobacterium tumefaciensADP-glucose pyrophosphorylase.
J. Biol. Chem., 294, 2019
|
|
5VWO
 
 | | Ornithine aminotransferase inactivated by (1R,3S,4S)-3-amino-4-fluorocyclopentane-1-carboxylic acid (FCP) | | Descriptor: | (1S,3S,4E)-3-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-4-iminocyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Mascarenhas, R, Liu, D, Le, H, Silverman, R. | | Deposit date: | 2017-05-22 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Selective Targeting by a Mechanism-Based Inactivator against Pyridoxal 5'-Phosphate-Dependent Enzymes: Mechanisms of Inactivation and Alternative Turnover.
Biochemistry, 56, 2017
|
|
5VWR
 
 | | E.coli Aspartate aminotransferase-(1R,3S,4S)-3-amino-4-fluorocyclopentane-1-carboxylic acid (FCP)-alpha-ketoglutarate | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-glutamic acid, Aspartate aminotransferase, GLYCEROL | | Authors: | Mascarenhas, R, Liu, D, Le, H, Silverman, R. | | Deposit date: | 2017-05-22 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Selective Targeting by a Mechanism-Based Inactivator against Pyridoxal 5'-Phosphate-Dependent Enzymes: Mechanisms of Inactivation and Alternative Turnover.
Biochemistry, 56, 2017
|
|
