7XDB
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron Spike protein in complex with BA7208 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7208 fab, ... | | Authors: | Liu, Z, Liu, S, Gao, Y.Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
7XDL
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7208 and BA7125 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7125 fab, ... | | Authors: | Liu, Z, Liu, S, Yuanzhu, G. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
6PRW
 
 | |
6PWH
 
 | |
7UTY
 
 | | First bromodomain of BRD4 liganded with compound 2c | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, prop-2-en-1-yl (5S)-1-ethyl-7-methyl-5-(4-methylphenyl)-2,4-dioxo-1,2,3,4,5,8-hexahydropyrido[2,3-d]pyrimidine-6-carboxylate | | Authors: | Schonbrunn, E, Chan, A. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1,4-Dihydropyridinebutyrolactone-derived ring-opened ester and amide analogs targeting BET bromodomains.
Arch Pharm, 355, 2022
|
|
8HRD
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Delta variant in complex with IMCAS74 Fab and W14 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS74 Fab heavy chain, IMCAS74 Fab light chain, ... | | Authors: | Zhao, R.C, Wu, L.L, Han, P. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6R2S
 
 | |
7Y3O
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, Light chain of BIOLS56, ... | | Authors: | Rao, X, Gao, F, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7Y3N
 
 | | Crystal structure of SARS-CoV receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, ... | | Authors: | Rao, X, Chai, Y, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4UV7
 
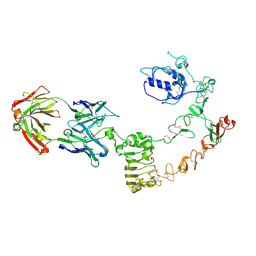 | | The complex structure of extracellular domain of EGFR and GC1118A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EPIDERMAL GROWTH FACTOR RECEPTOR, ... | | Authors: | Yoo, J.H, Cho, H.S. | | Deposit date: | 2014-08-05 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Gc1118, an Anti-Egfr Antibody with a Distinct Binding Epitope and Superior Inhibitory Activity Against High-Affinity Egfr Ligands.
Mol.Cancer Ther., 15, 2016
|
|
4HR3
 
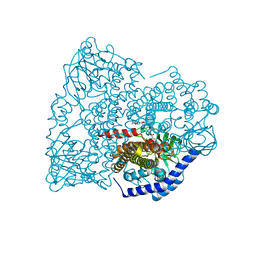 | |
4I1Y
 
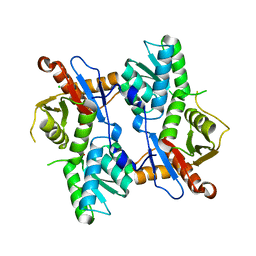 | |
4IV6
 
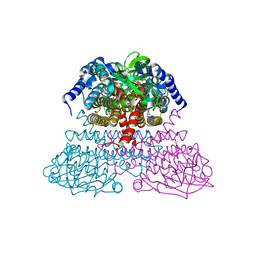 | |
4IZ9
 
 | |
4J3J
 
 | | Crystal Structure of DPP-IV with Compound C3 | | Descriptor: | Dipeptidyl peptidase 4, N-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butyl]-6-(trifluoromethyl)-3,4-dihydropyrrolo[1,2-a]pyrazine-2(1H)-carboxamide | | Authors: | Xiong, B, Zhu, L.R, Chen, D.Q, Zhao, Y.L, Jiang, F, Shen, J.K. | | Deposit date: | 2013-02-05 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and synthesis of 4-(2,4,5-trifluorophenyl)butane-1,3-diamines as dipeptidyl peptidase IV inhibitors
Chemmedchem, 8, 2013
|
|
4J5I
 
 | |
4JLV
 
 | | Crystal structure of the chimerical protein CapA1B1 in complex with ADP-Mg | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, C-terminal fragment of Membrane protein CapA1, Putative uncharacterized protein capB1, ... | | Authors: | Gruszczyk, J, Olivares-Illana, V, Nourikyan, J, Fleurie, A, Bechet, E, Aumont-Nicaise, M, Gueguen-Chaignon, V, Morera, S, Grangeasse, C, Nessler, S. | | Deposit date: | 2013-03-13 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparative analysis of the Tyr-kinases CapB1 and CapB2 fused to their cognate modulators CapA1 and CapA2 from Staphylococcus aureus
Plos One, 8, 2013
|
|
4HDT
 
 | |
4IJN
 
 | |
1GYQ
 
 | |
4JMP
 
 | | Crystal structure of the chimerical protein CapA2B2 | | Descriptor: | C-terminal fragment of CapA, Protein tyrosine kinase | | Authors: | Olivares-Illana, V, Morera, S, Grangeasse, C, Nessler, S. | | Deposit date: | 2013-03-14 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Comparative analysis of the Tyr-kinases CapB1 and CapB2 fused to their cognate modulators CapA1 and CapA2 from Staphylococcus aureus
Plos One, 8, 2013
|
|
4KAM
 
 | |
4HS3
 
 | | Crystal structure of H-2Kb with a disulfide stabilized F pocket in complex with the LCMV derived peptide GP34 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-2-microglobulin, Envelope glycoprotein, ... | | Authors: | Uchtenhagen, H, Boulanger, B, Hein, Z, Abualrous, E.T, Zacharias, M, Werner, J, Elliott, T, Springer, S, Achour, A. | | Deposit date: | 2012-10-29 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Peptide-independent stabilization of MHC class I molecules breaches cellular quality control.
J.Cell.Sci., 127, 2014
|
|
3V4O
 
 | |
3CWB
 
 | | Chicken Cytochrome BC1 Complex inhibited by an iodinated analogue of the polyketide Crocacin-D | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, AZIDE ION, CARDIOLIPIN, ... | | Authors: | Huang, L, Cromartie, T, Viner, R, Crowley, P.J, Berry, E.A. | | Deposit date: | 2008-04-21 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | The role of molecular modeling in the design of analogues of the fungicidal natural products crocacins A and D.
Bioorg.Med.Chem., 16, 2008
|
|
