4R7E
 
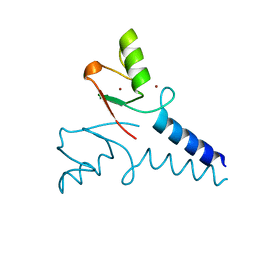 | | Structure of Bre1 RING domain | | Descriptor: | E3 ubiquitin-protein ligase BRE1, ZINC ION | | Authors: | Kumar, P, Wolberger, C. | | Deposit date: | 2014-08-27 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Structure of the yeast Bre1 RING domain.
Proteins, 83, 2015
|
|
2ECW
 
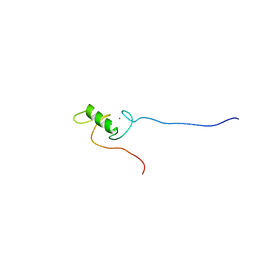 | | Solution structure of the Zinc finger, C3HC4 type (RING finger) domain Tripartite motif protein 30 | | Descriptor: | Tripartite motif-containing protein 30, ZINC ION | | Authors: | Abe, H, Miyamoto, K, Tochio, N, Yoneyama, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-14 | | Release date: | 2008-02-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Zinc finger, C3HC4 type (RING finger) domain Tripartite motif protein 30
To be Published
|
|
5YUF
 
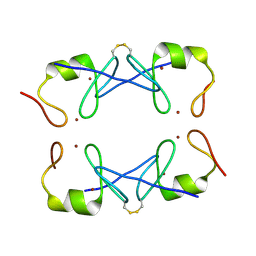 | | Crystal Structure of PML RING tetramer | | Descriptor: | Protein PML, ZINC ION | | Authors: | Wang, P, Benhend, S, Wu, H, Breitenbach, V, Zhen, T, Jollivet, F, Peres, L, Li, Y, Chen, S, Chen, Z, de THE, H, Meng, G. | | Deposit date: | 2017-11-22 | | Release date: | 2018-04-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | RING tetramerization is required for nuclear body biogenesis and PML sumoylation.
Nat Commun, 9, 2018
|
|
1CHC
 
 | |
2LDR
 
 | |
2MWX
 
 | | The RING Domain of human Promyelocytic Leukemia Protein (PML) | | Descriptor: | Protein PML, ZINC ION | | Authors: | Huang, S.Y, Chang, C.F, Fan, P.J, Guntert, P, Shih, H.M, Huang, T.H. | | Deposit date: | 2014-12-02 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The RING domain of human promyelocytic leukemia protein (PML).
J.Biomol.Nmr, 61, 2015
|
|
2K4D
 
 | | E2-c-Cbl recognition is necessary but not sufficient for ubiquitination activity | | Descriptor: | E3 ubiquitin-protein ligase CBL, ZINC ION | | Authors: | Huang, A, De Jong, R.N, Wienk, H, Winkler, S.G, Timmers, H.T.M, Boelens, R. | | Deposit date: | 2008-06-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | E2-c-Cbl recognition is necessary but not sufficient for ubiquitination activity
J.Mol.Biol., 385, 2009
|
|
5H7R
 
 | |
1BOR
 
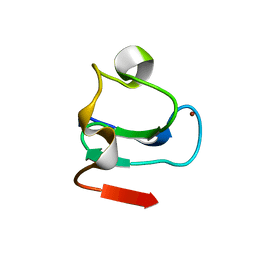 | | TRANSCRIPTION FACTOR PML, A PROTO-ONCOPROTEIN, NMR, 1 REPRESENTATIVE STRUCTURE AT PH 7.5, 30 C, IN THE PRESENCE OF ZINC | | Descriptor: | TRANSCRIPTION FACTOR PML, ZINC ION | | Authors: | Borden, K.L.B, Freemont, P.S. | | Deposit date: | 1995-09-27 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML.
EMBO J., 14, 1995
|
|
5TRB
 
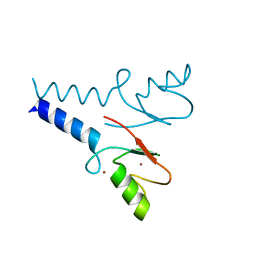 | | Crystal structure of the RNF20 RING domain | | Descriptor: | E3 ubiquitin-protein ligase BRE1A, ZINC ION | | Authors: | Foglizzo, M, Middleton, A.J, Day, C.L. | | Deposit date: | 2016-10-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Function of the RING Domains of RNF20 and RNF40, Dimeric E3 Ligases that Monoubiquitylate Histone H2B.
J.Mol.Biol., 428, 2016
|
|
7OD1
 
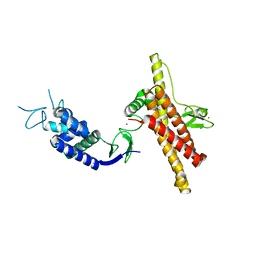 | |
7LYB
 
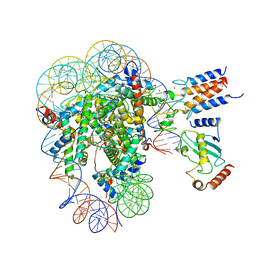 | | Cryo-EM structure of the human nucleosome core particle in complex with BRCA1-BARD1-UbcH5c | | Descriptor: | BRCA1-associated RING domain protein 1, DNA (146-MER), DNA (147-MER), ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, D, Mer, E, Mer, G. | | Deposit date: | 2021-03-06 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Mechanisms of BRCA1-BARD1 nucleosome recognition and ubiquitylation.
Nature, 596, 2021
|
|
6FGA
 
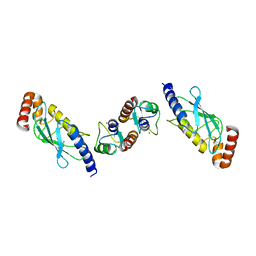 | | Crystal structure of TRIM21 E3 ligase, RING domain in complex with its cognate E2 conjugating enzyme UBE2E1 | | Descriptor: | E3 ubiquitin-protein ligase TRIM21, GLYCEROL, Ubiquitin-conjugating enzyme E2 E1, ... | | Authors: | Anandapadamanaban, M, Moche, M, Sunnerhagen, M. | | Deposit date: | 2018-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | E3 ubiquitin-protein ligase TRIM21-mediated lysine capture by UBE2E1 reveals substrate-targeting mode of a ubiquitin-conjugating E2.
J.Biol.Chem., 294, 2019
|
|
6V0V
 
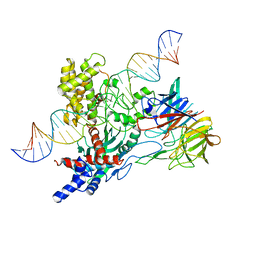 | | Cryo-EM structure of mouse WT RAG1/2 NFC complex (DNA0) | | Descriptor: | CALCIUM ION, DNA (30-MER), V(D)J recombination-activating protein 1, ... | | Authors: | Chen, X, Yang, W, Gellert, M. | | Deposit date: | 2019-11-19 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3VGO
 
 | |
7JZV
 
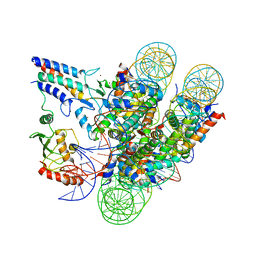 | | Cryo-EM structure of the BRCA1-UbcH5c/BARD1 E3-E2 module bound to a nucleosome | | Descriptor: | BRCA1,Ubiquitin-conjugating enzyme E2 D3, BRCA1-associated RING domain protein 1, Histone H2A type 2-A, ... | | Authors: | Witus, S.R, Burrell, A.L, Hansen, J.M, Farrell, D.P, Dimaio, F, Kollman, J.M, Klevit, R.E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | BRCA1/BARD1 site-specific ubiquitylation of nucleosomal H2A is directed by BARD1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1JM7
 
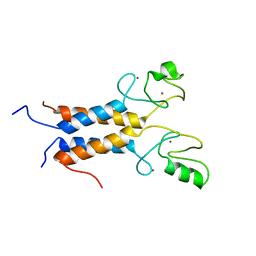 | | Solution structure of the BRCA1/BARD1 RING-domain heterodimer | | Descriptor: | BRCA1-ASSOCIATED RING DOMAIN PROTEIN 1, BREAST CANCER TYPE 1 SUSCEPTIBILITY PROTEIN, ZINC ION | | Authors: | Brzovic, P.S, Rajagopal, P, Hoyt, D.W, King, M.-C, Klevit, R.E. | | Deposit date: | 2001-07-17 | | Release date: | 2001-10-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of a BRCA1-BARD1 heterodimeric RING-RING complex.
Nat.Struct.Biol., 8, 2001
|
|
1RMD
 
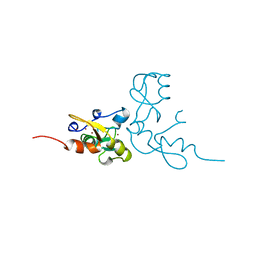 | | RAG1 DIMERIZATION DOMAIN | | Descriptor: | RAG1, ZINC ION | | Authors: | Bellon, S.F, Rodgers, K.K, Schatz, D.G, Coleman, J.E, Steitz, T.A. | | Deposit date: | 1997-01-10 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster.
Nat.Struct.Biol., 4, 1997
|
|
6L8O
 
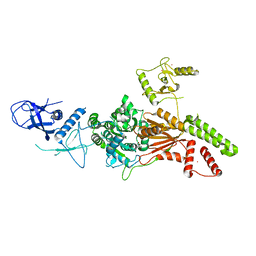 | | Crystal structure of the K. lactis Rad5 (Hg-derivative) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA repair protein RAD5, MERCURY (II) ION | | Authors: | Shen, M, Xiang, S. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
8A58
 
 | |
6L8N
 
 | | Crystal structure of the K. lactis Rad5 | | Descriptor: | DNA repair protein RAD5, ZINC ION | | Authors: | Shen, M, Xiang, S. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
2Y1M
 
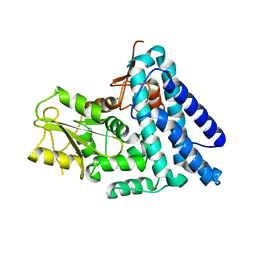 | | Structure of native c-Cbl | | Descriptor: | CALCIUM ION, E3 UBIQUITIN-PROTEIN LIGASE, ZINC ION | | Authors: | Dou, H, Sibbet, G.J, Huang, D.T. | | Deposit date: | 2010-12-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural Basis for Autoinhibition and Phosphorylation-Dependent Activation of C-Cbl.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5O76
 
 | | Structure of phosphoY371 c-CBL in complex with ZAP70-peptide and UbV.pCBL ubiquitin variant | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase CBL, Tyrosine protein kinase ZAP70 peptide, ... | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2017-06-08 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | A General Strategy for Discovery of Inhibitors and Activators of RING and U-box E3 Ligases with Ubiquitin Variants.
Mol. Cell, 68, 2017
|
|
4KBL
 
 | | Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, ZINC ION | | Authors: | Duda, D.M, Olszewski, J.L, Schulman, B.A. | | Deposit date: | 2013-04-23 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of HHARI, a RING-IBR-RING Ubiquitin Ligase: Autoinhibition of an Ariadne-Family E3 and Insights into Ligation Mechanism.
Structure, 21, 2013
|
|
8GCY
 
 | | Co-crystal structure of CBL-B in complex with N-Aryl isoindolin-1-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Kimani, S, Zeng, H, Dong, A, Li, Y, Santhakumar, V, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The co-crystal structure of Cbl-b and a small-molecule inhibitor reveals the mechanism of Cbl-b inhibition.
Commun Biol, 6, 2023
|
|
