1QYT
 
 | | Solution structure of fragment (25-35) of beta amyloid peptide in SDS micellar solution | | Descriptor: | 11-mer peptide from Amyloid beta A4 protein | | Authors: | D'Ursi, A.M, Armenante, M.R, Guerrini, R, Salvadori, S, Sorrentino, G, Picone, D. | | Deposit date: | 2003-09-12 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of amyloid beta-peptide (25-35) in different media
J.Med.Chem., 47, 2004
|
|
1QCM
 
 | | AMYLOID BETA PEPTIDE (25-35), NMR, 20 STRUCTURES | | Descriptor: | AMYLOID BETA PEPTIDE | | Authors: | Kohno, T, Kobayashi, K, Maeda, T, Sato, K, Takashima, A. | | Deposit date: | 1996-07-19 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structures of the amyloid beta peptide (25-35) in membrane-mimicking environment.
Biochemistry, 35, 1996
|
|
8SEK
 
 | |
8SEL
 
 | |
8SEJ
 
 | |
3IFN
 
 | | X-ray structure of amyloid beta peptide:antibody (Abeta1-40:12A11) complex | | Descriptor: | 12A11 FAB antibody heavy chain, 12A11 FAB antibody light chain, Amyloid beta A4 protein | | Authors: | Weis, W.I, Feinberg, H, Basi, G.S, Schenk, D. | | Deposit date: | 2009-07-24 | | Release date: | 2009-11-17 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural correlates of antibodies associated with acute reversal of amyloid beta-related behavioral deficits in a mouse model of Alzheimer disease.
J.Biol.Chem., 285, 2010
|
|
3IFO
 
 | | X-ray structure of amyloid beta peptide:antibody (Abeta1-7:10D5) complex | | Descriptor: | 10D5 FAB antibody heavy chain, 10D5 FAB antibody light chain, Amyloid beta A4 protein | | Authors: | Weis, W.I, Feinberg, H, Basi, G.S, Schenk, D. | | Deposit date: | 2009-07-24 | | Release date: | 2009-11-17 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural correlates of antibodies associated with acute reversal of amyloid beta-related behavioral deficits in a mouse model of Alzheimer disease.
J.Biol.Chem., 285, 2010
|
|
3IFP
 
 | | X-ray structure of amyloid beta peptide:antibody (Abeta1-7:12B4) complex | | Descriptor: | 12B4 FAB antibody heavy chain, 12B4 FAB antibody light chain, Amyloid beta A4 protein | | Authors: | Weis, W.I, Feinberg, H, Basi, G.S, Schenk, D. | | Deposit date: | 2009-07-24 | | Release date: | 2009-11-17 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural correlates of antibodies associated with acute reversal of amyloid beta-related behavioral deficits in a mouse model of Alzheimer disease.
J.Biol.Chem., 285, 2010
|
|
3IFL
 
 | | X-ray structure of amyloid beta peptide:antibody (Abeta1-7:12A11) complex | | Descriptor: | 12A11 FAB antibody heavy chain, 12A11 FAB antibody light chain, Amyloid beta A4 protein | | Authors: | Weis, W.I, Feinberg, H, Basi, G.S, Schenk, D. | | Deposit date: | 2009-07-24 | | Release date: | 2009-11-17 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural correlates of antibodies associated with acute reversal of amyloid beta-related behavioral deficits in a mouse model of Alzheimer disease.
J.Biol.Chem., 285, 2010
|
|
3JTI
 
 | | Crystal structure of the complex formed between Phospholipase A2 with beta-amyloid fragment, Lys-Gly-Ala-Ile-Ile-Gly-Leu-Met at 1.8 A resolution | | Descriptor: | CALCIUM ION, Phospholipase A2 isoform 3, octapeptide from Amyloid beta A4 protein | | Authors: | Pandey, N, Mirza, Z, Vikram, G, Singh, N, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-09-12 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex formed between Phospholipase A2 with beta-amyloid fragment, Lys-Gly-Ala-Ile-Ile-Gly-Leu-Met at 1.8 A resolution
To be Published
|
|
3NYJ
 
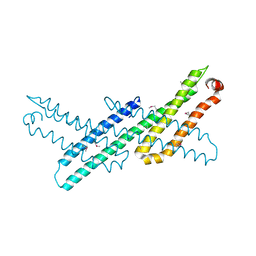 | | Crystal Structure Analysis of APP E2 domain | | Descriptor: | Amyloid beta A4 protein, OSMIUM ION | | Authors: | Ha, Y, Hu, J, Lee, S, Liu, X. | | Deposit date: | 2010-07-15 | | Release date: | 2011-06-01 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The E2 Domains of APP and APLP1 Share a Conserved Mode of Dimerization.
Biochemistry, 50, 2011
|
|
3OW9
 
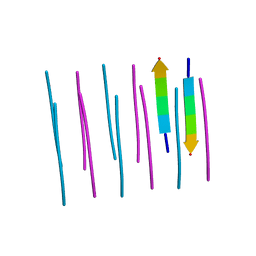 | |
3OVJ
 
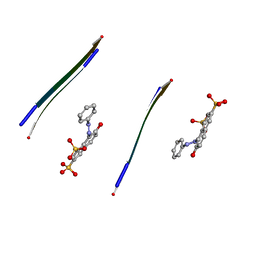 | |
2BEG
 
 | | 3D Structure of Alzheimer's Abeta(1-42) fibrils | | Descriptor: | Amyloid beta A4 protein | | Authors: | Luhrs, T, Ritter, C, Adrian, M, Riek-Loher, D, Bohrmann, B, Dobeli, H, Schubert, D, Riek, R. | | Deposit date: | 2005-10-24 | | Release date: | 2005-11-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 3D structure of Alzheimer's amyloid-{beta}(1-42) fibrils.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
8AZT
 
 | | Type II amyloid-beta 42 filaments from high-spin supernatants of aqueous extracts from Alzheimer's disease brains | ABeta42 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Yang, Y, Stern, M.A, Meunier, L.A, Liu, W, Cai, Y.Q, Ericsson, M, Liu, L, Selkoe, J.D, Goedert, M, Scheres, H.W.S. | | Deposit date: | 2022-09-06 | | Release date: | 2022-11-02 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Abundant A beta fibrils in ultracentrifugal supernatants of aqueous extracts from Alzheimer's disease brains.
Neuron, 111, 2023
|
|
8AZS
 
 | | Type I amyloid-beta 42 filaments from high-spin supernatants of aqueous extracts from Alzheimer's disease brains | ABeta42 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Yang, Y, Stern, M.A, Meunier, L.A, Liu, W, Cai, Y.Q, Ericsson, M, Liu, L, Selkoe, J.D, Goedert, M, Scheres, H.W.S. | | Deposit date: | 2022-09-06 | | Release date: | 2022-11-02 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Abundant A beta fibrils in ultracentrifugal supernatants of aqueous extracts from Alzheimer's disease brains.
Neuron, 111, 2023
|
|
2BP4
 
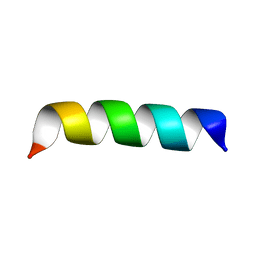 | | Zinc-binding domain of Alzheimer's disease amyloid beta-peptide in TFE-water (80-20) solution | | Descriptor: | AMYLOID BETA A4 PROTEIN | | Authors: | Zirah, S, Kozin, S.A, Mazur, A.K, Blond, A, Cheminant, M, Segalas-Milazzo, I, Debey, P, Rebuffat, S. | | Deposit date: | 2005-04-18 | | Release date: | 2005-04-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Changes of Region 1-16 of the Alzheimer Disease Amyloid Beta-Peptide Upon Zinc Binding and in Vitro Aging.
J.Biol.Chem., 281, 2006
|
|
8BG0
 
 | | Amyloid-beta tetrameric filaments with the Arctic mutation (E22G) from Alzheimer's disease brains | ABeta40 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Yang, Y, Zhang, W.J, Murzin, A.G, Schweighauser, M, Huang, M, Lovestam, S.K.A, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Ghetti, B, Graff, C, Kumar, A, Nordber, A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2022-10-27 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Cryo-EM structures of amyloid-beta filaments with the Arctic mutation (E22G) from human and mouse brains.
Acta Neuropathol, 145, 2023
|
|
8BFZ
 
 | | Amyloid-beta 42 filaments extracted from the human brain with Arctic mutation (E22G) of Alzheimer's disease | ABeta42 | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Yang, Y, Zhang, W.J, Murzin, A.G, Schweighauser, M, Huang, M, Lovestam, S.K.A, Peak-Chew, S.Y, Macdonald, J, Lavenir, I, Ghetti, B, Graff, C, Kumar, A, Nordberg, A, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2022-10-27 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of amyloid-beta filaments with the Arctic mutation (E22G) from human and mouse brains.
Acta Neuropathol, 145, 2023
|
|
8BFA
 
 | | Sarkosyl-extracted AppNL-G-F Abeta42 fibril structure | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Wilkinson, M, Leistner, C, Burgess, A, Goodfellow, S, Deuchars, S, Ranson, N.A, Radford, S.E, Frank, R.A.W. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The in-tissue molecular architecture of beta-amyloid pathology in the mammalian brain.
Nat Commun, 14, 2023
|
|
8BFB
 
 | | Sarkosyl-extracted AppNL-G-F Abeta42 fibril structure (Methoxy-X04-labelled mice) | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Wilkinson, M, Leistner, C, Burgess, A, Goodfellow, S, Deuchars, S, Ranson, N.A, Radford, S.E, Frank, R.A.W. | | Deposit date: | 2022-10-24 | | Release date: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The in-tissue molecular architecture of beta-amyloid pathology in the mammalian brain.
Nat Commun, 14, 2023
|
|
2FK3
 
 | |
2FJZ
 
 | |
2FKL
 
 | |
2FK1
 
 | |
