8I5C
 
 | | Crystal structure of a TCR in complex with HLA-A*11:01 bound to KRAS peptide (VVGAVGVGK) | | Descriptor: | Beta-2-microglobulin, MHC class I antigen (Fragment), TCR alpha chain, ... | | Authors: | Lu, D, Chen, Y, Jiang, M, Tan, S.G, Chai, Y, Gao, G.F. | | Deposit date: | 2023-01-24 | | Release date: | 2023-08-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Crystal structure of a TCR in complex with HLA-A*11:01 bound to KRAS peptide (VVGAVGVGK)
Nat Commun, 2023
|
|
6E6F
 
 | | KRAS G13D bound to GppNHp (K13GNP) | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Johnson, C.W, Mattos, C. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Isoform-Specific Destabilization of the Active Site Reveals a Molecular Mechanism of Intrinsic Activation of KRas G13D.
Cell Rep, 28, 2019
|
|
7KFZ
 
 | | Structure of a ternary KRas(G13D)-SOS complex | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Liu, C, Moghadamchargari, Z, Laganowsky, A, Zhao, M. | | Deposit date: | 2020-10-15 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Molecular assemblies of the catalytic domain of SOS with KRas and oncogenic mutants.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8QU8
 
 | | PROTAC-mediated complex of KRAS with VHL/Elongin-B/Elongin-C/Cullin-2/Rbx1 | | Descriptor: | (2S,4R)-1-[(2S)-2-[4-[4-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]butoxy]-1,2,3-triazol-1-yl]-3-methyl-butanoyl]-N-[(1R)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-2-oxidanyl-ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Fischer, G, Peter, D, Arce-Solano, S. | | Deposit date: | 2023-10-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Targeting cancer with small-molecule pan-KRAS degraders.
Science, 385, 2024
|
|
7W5R
 
 | | KRAS G12V and peptide complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, LEU-TYR-ASP-VAL-ALA, ... | | Authors: | Kim, H.J, Han, C.W, Jang, S.B. | | Deposit date: | 2021-11-30 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.87 Å) | | Cite: | Structural basis of the oncogenic KRAS mutant and GJ101 complex.
Biochem.Biophys.Res.Commun., 641, 2023
|
|
8DGT
 
 | | Cryo-EM structure of a RAS/RAF complex (state 2) | | Descriptor: | 14-3-3 protein zeta, 5-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Eck, M.J, Jeon, H, Park, E, Rawson, S. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of a RAS/RAF recruitment complex.
Nat Commun, 14, 2023
|
|
8DGS
 
 | | Cryo-EM structure of a RAS/RAF complex (state 1) | | Descriptor: | 14-3-3 protein zeta, 5-[(2-fluoro-4-iodophenyl)amino]-N-(2-hydroxyethoxy)imidazo[1,5-a]pyridine-6-carboxamide, Dual specificity mitogen-activated protein kinase kinase 1, ... | | Authors: | Eck, M.J, Jeon, H, Park, E, Rawson, S. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of a RAS/RAF recruitment complex.
Nat Commun, 14, 2023
|
|
7RSE
 
 | | NMR-driven structure of the KRAS4B-G12D "alpha-beta" dimer on a lipid bilayer nanodisc | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
7RSC
 
 | | NMR-driven structure of the KRAS4B-G12D "alpha-alpha" dimer on a lipid bilayer nanodisc | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
7MQU
 
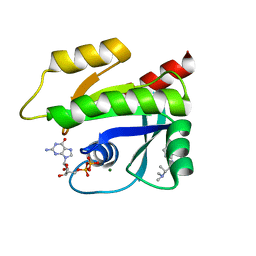 | | The haddock model of GDP KRas in complex with promethazine using NMR chemical shift perturbations | | Descriptor: | (2R)-N,N-dimethyl-1-(10H-phenothiazin-10-yl)propan-2-amine, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, X, Gorfe, A.A, Putkey, J.P. | | Deposit date: | 2021-05-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Antipsychotic phenothiazine drugs bind to KRAS in vitro.
J.Biomol.Nmr, 75, 2021
|
|
6V5L
 
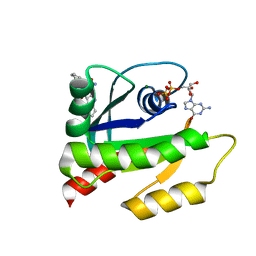 | | The HADDOCK structure model of GDP KRas in complex with its allosteric inhibitor E22 | | Descriptor: | (2R)-2-[2-(1H-indole-3-carbonyl)hydrazinyl]-2-phenylacetamide, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, X, Gupta, A.K, Prakash, P, Putkey, J.P, Gorfe, A.A. | | Deposit date: | 2019-12-04 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multi target ensemble based virtual screening yields novel allosteric KRAS inhibitors at high success rate
Chemical Biology & Drug Design, 94, 2019
|
|
6PTS
 
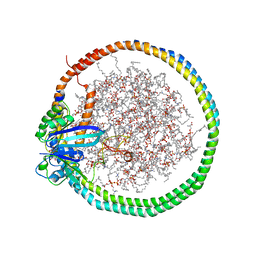 | | NMR data-driven model of KRas-GMPPNP:RBD-CRD complex tethered to a nanodisc (state A) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Lee, K, Gasmi-Seabrook, G, Ikura, M, Marshall, C.B. | | Deposit date: | 2019-07-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multivalent assembly of KRAS with the RAS-binding and cysteine-rich domains of CRAF on the membrane.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PTW
 
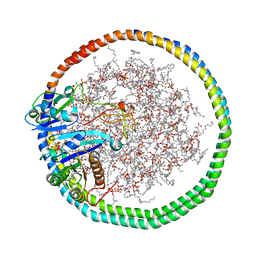 | | NMR data-driven model of KRas-GMPPNP:RBD-CRD complex tethered to a nanodisc (state B) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Lee, K, Gasmi-Seabrook, G, Ikura, M, Marshall, C.B. | | Deposit date: | 2019-07-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multivalent assembly of KRAS with the RAS-binding and cysteine-rich domains of CRAF on the membrane.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6W4F
 
 | | NMR-driven structure of KRAS4B-GDP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
2MSD
 
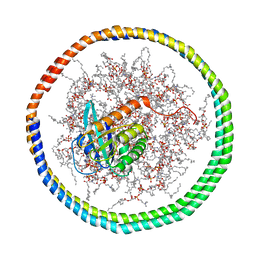 | | NMR data-driven model of GTPase KRas-GNP tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6W4E
 
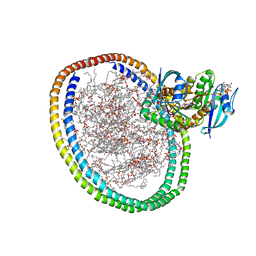 | | NMR-driven structure of KRAS4B-GTP homodimer on a lipid bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, ... | | Authors: | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | Deposit date: | 2020-03-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6CC9
 
 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | Descriptor: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-06 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
7KYZ
 
 | |
8PI0
 
 | |
8PIY
 
 | |
7LGI
 
 | | The haddock model of GDP KRas in complex with promazine using chemical shift perturbations and intermolecular NOEs | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, X, Gorfe, A.A, Putkey, J.A. | | Deposit date: | 2021-01-20 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Antipsychotic phenothiazine drugs bind to KRAS in vitro.
J.Biomol.Nmr, 75, 2021
|
|
6CCX
 
 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | Descriptor: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
6CCH
 
 | | NMR data-driven model of GTPase KRas-GMPPNP tethered to a nanodisc (E3 state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
2MSE
 
 | | NMR data-driven model of GTPase KRas-GNP:ARafRBD complex tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MSC
 
 | | NMR data-driven model of GTPase KRas-GDP tethered to a lipid-bilayer nanodisc | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
