8T3O
 
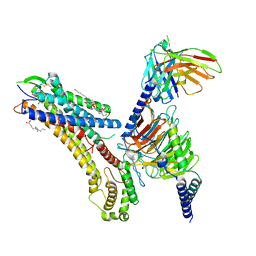 | | Cryo-EM structure of the TUG-891 bound FFA4-Gq complex | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8T3V
 
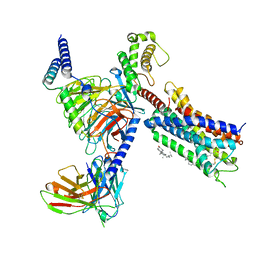 | | Cryo-EM structure of the DHA bound FFA1-Gq complex | | Descriptor: | CHOLESTEROL, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 1, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
6AG0
 
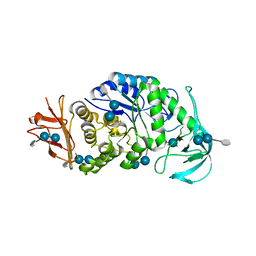 | | The X-ray Crystallographic Structure of Maltooligosaccharide-forming Amylase from Bacillus stearothermophilus STB04 | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-amylase, CALCIUM ION | | Authors: | Li, Z.F, Li, Y.L, Ban, X.F, Zhang, C.Y, Jin, T.C, Xie, X.F, Gu, Z.B, Li, C.M. | | Deposit date: | 2018-08-09 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a maltooligosaccharide-forming amylase from Bacillus stearothermophilus STB04.
Int.J.Biol.Macromol., 138, 2019
|
|
5FBH
 
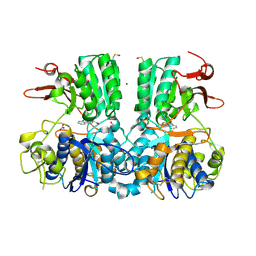 | | Crystal structure of the extracellular domain of human calcium sensing receptor with bound Gd3+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Zhang, T, Zhang, C, Miller, C.L, Zou, J, Moremen, K.W, Brown, E.M, Yang, J.J, Hu, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist.
Sci Adv, 2, 2016
|
|
5FBK
 
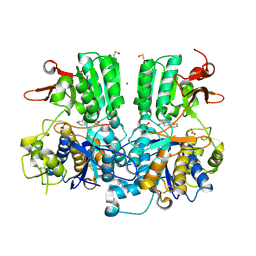 | | Crystal structure of the extracellular domain of human calcium sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Zhang, T, Zhang, C, Miller, C.L, Zou, J, Moremen, K.W, Brown, E.M, Yang, J.J, Hu, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist.
Sci Adv, 2, 2016
|
|
8T3Q
 
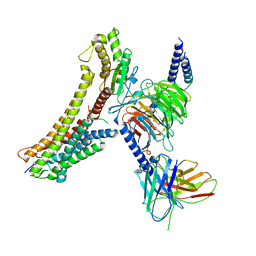 | | Cryo-EM structure of the DHA bound FFA4-Gq complex | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
3PFT
 
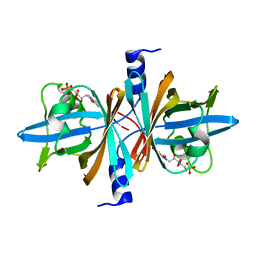 | | Crystal Structure of Untagged C54A Mutant Flavin Reductase (DszD) in Complex with FMN From Mycobacterium goodii | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin reductase | | Authors: | Li, Q, Xu, P, Ma, C, Gu, L, Liu, X, Zhang, C, Li, N, Su, J, Li, B, Liu, S. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | The flavin reductase DSZD from a desulfurizing mycobacterium goodii strain: systemic manipulation and investigation based on the crystal structure
To be Published
|
|
4HQX
 
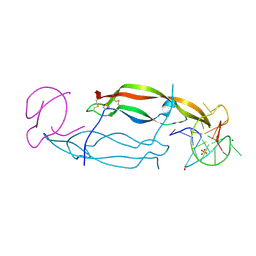 | | CRYSTAL STRUCTURE OF HUMAN PDGF-BB IN COMPLEX WITH A Modified nucleotide aptamer (SOMAmer SL4) | | Descriptor: | MAGNESIUM ION, Platelet-derived growth factor subunit B, SODIUM ION, ... | | Authors: | Davies, D.R, Edwards, T.E, Janjic, N, Gelinas, A.D, Zhang, C, Jarvis, T.C. | | Deposit date: | 2012-10-26 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4HQU
 
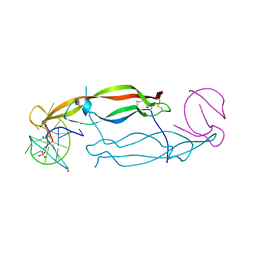 | | Crystal structure of human PDGF-BB in complex with a modified nucleotide aptamer (SOMAmer SL5) | | Descriptor: | MAGNESIUM ION, Platelet-derived growth factor subunit B, SODIUM ION, ... | | Authors: | Davies, D.R, Edwards, T.E, Janjic, N, Gelinas, A.D, Zhang, C, Jarvis, T.C. | | Deposit date: | 2012-10-26 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2WEI
 
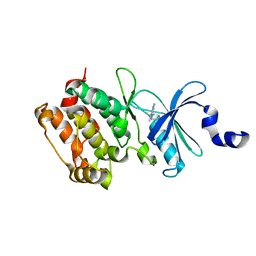 | | Crystal structure of the kinase domain of Cryptosporidium parvum calcium dependent protein kinase in complex with 3-MB-PP1 | | Descriptor: | 1-tert-butyl-3-(3-methylbenzyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALMODULIN-DOMAIN PROTEIN KINASE 1, PUTATIVE | | Authors: | Roos, A.K, King, O, Chaikuad, A, Zhang, C, Shokat, K.M, Wernimont, A.K, Artz, J.D, Lin, L, MacKenzie, F.I, Finerty, P.J, Vedadi, M, Schapira, M, Indarte, M, Kozieradzki, I, Pike, A.C.W, Fedorov, O, Doyle, D, Muniz, J, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Heightman, T, Hui, R. | | Deposit date: | 2009-03-31 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Cryptosporidium Parvum Kinome.
Bmc Genomics, 12, 2011
|
|
7VL8
 
 | | Cryo-EM structure of the Apo CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
8G59
 
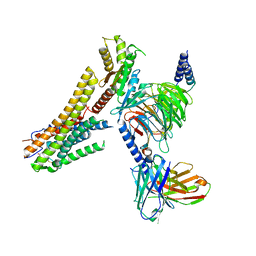 | | Cryo-EM structure of the TUG891 bound GPR120-Giq complex | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-08 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID6
 
 | | Cryo-EM structure of the oleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID9
 
 | | Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi complex | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID4
 
 | | Cryo-EM structure of the linoleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID3
 
 | | Cryo-EM structure of the 9-hydroxystearic acid bound GPR120-Gi complex | | Descriptor: | 9-Hydroxyoctadecanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID8
 
 | | Cryo-EM structure of the TUG891 bound GPR120-Gi complex | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
6CAD
 
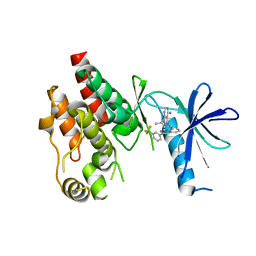 | | Crystal structure of RAF kinase domain bound to the inhibitor 2a | | Descriptor: | 1-(propan-2-yl)-3-({3-[3-(trifluoromethyl)phenyl]isoquinolin-8-yl}ethynyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Serine/threonine-protein kinase B-raf | | Authors: | Maisonneuve, P, Kurinov, I, Assadieskandar, A, Yu, C, Liu, X, Chen, Y.-C, Prakash, G.K.S, Zhang, C, SIcheri, F. | | Deposit date: | 2018-01-30 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Effects of rigidity on the selectivity of protein kinase inhibitors.
Eur J Med Chem, 146, 2018
|
|
4IAR
 
 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with ergotamine (PSI Community Target) | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Ergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
4IAQ
 
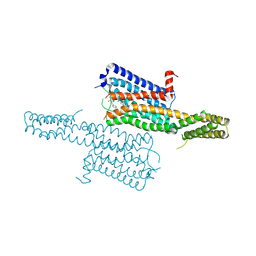 | | Crystal structure of the chimeric protein of 5-HT1B-BRIL in complex with dihydroergotamine (PSI Community Target) | | Descriptor: | Chimera protein of human 5-hydroxytryptamine receptor 1B and E. Coli soluble cytochrome b562, Dihydroergotamine | | Authors: | Wang, C, Jiang, Y, Ma, J, Wu, H, Wacker, D, Katritch, V, Han, G.W, Liu, W, Huang, X, Vardy, E, McCorvy, J.D, Gao, X, Zhou, E.X, Melcher, K, Zhang, C, Bai, F, Yang, H, Yang, L, Jiang, H, Roth, B.L, Cherezov, V, Stevens, R.C, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for molecular recognition at serotonin receptors.
Science, 340, 2013
|
|
8JUA
 
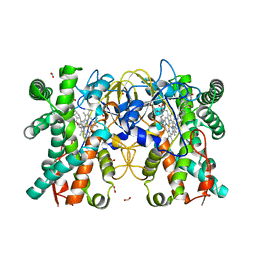 | | Multifunctional cytochrome P450 enzyme IkaD from Streptomyces sp. ZJ306, in complex with epoxyikarugamycin | | Descriptor: | (1Z,3E,5S,7R,8R,10R,11R,12S,13R,15S,16R,17S,19Z,26S)-11-ethyl-2-hydroxy-10-methyl-22,27-diaza-14 oxahexacyclo[24.2.1.05,17.07,16.013,15.08,12]nonacosa-1(2),3,19-triene-21,28,29-trione, Cytochrome P450, FORMIC ACID, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.00001121 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7VLA
 
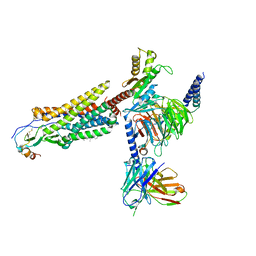 | | Cryo-EM structure of the CCL15(27-92) bound CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CCL15(27-92), CHOLESTEROL, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7VL9
 
 | | Cryo-EM structure of the CCL15(26-92) bound CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CCL15(26-92), CHOLESTEROL, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7VWP
 
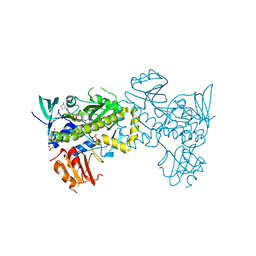 | | Structure of the flavin-dependent monooxygenase FlsO1 from the biosynthesis of fluostatinsin | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FlsO1, PHOSPHATE ION, ... | | Authors: | Zhang, Y, Yang, C, Zhang, L, Zhang, C. | | Deposit date: | 2021-11-11 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and structural insights of multifunctional flavin-dependent monooxygenase FlsO1-catalyzed unexpected xanthone formation
Nat Commun, 13, 2022
|
|
3PDS
 
 | | Irreversible Agonist-Beta2 Adrenoceptor Complex | | Descriptor: | 8-hydroxy-5-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(3-sulfanylpropoxy)phenyl]ethyl}amino)ethyl]quinolin-2(1H)-one, CHOLESTEROL, Fusion protein Beta-2 adrenergic receptor/Lysozyme, ... | | Authors: | Rosenbaum, D.M, Zhang, C, Lyons, J.A, Holl, R, Aragao, D, Arlow, D.H, Rasmussen, S.G.F, Choi, H.-J, DeVree, B.T, Sunahara, R.K, Chae, P.S, Gellman, S.H, Dror, R.O, Shaw, D.E, Weis, W.I, Caffrey, M, Gmeiner, P, Kobilka, B.K. | | Deposit date: | 2010-10-24 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and function of an irreversible agonist-beta(2) adrenoceptor complex
Nature, 469, 2011
|
|
