5TXK
 
 | | CRYSTAL STRUCTURE OF USP35 C450S IN COMPLEX WITH UBIQUITIN | | Descriptor: | 1,2-ETHANEDIOL, Polyubiquitin-B, SULFATE ION, ... | | Authors: | Bader, G, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2016-11-17 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Expansion of DUB functionality generated by alternative isoforms - USP35, a case study.
J. Cell. Sci., 131, 2018
|
|
3C7Q
 
 | | Structure of VEGFR2 kinase domain in complex with BIBF1120 | | Descriptor: | SULFATE ION, Vascular endothelial growth factor receptor 2, methyl (3Z)-3-{[(4-{methyl[(4-methylpiperazin-1-yl)acetyl]amino}phenyl)amino](phenyl)methylidene}-2-oxo-2,3-dihydro-1H-indole-6-carboxylate | | Authors: | Hilberg, F, Roth, G.J, Krssak, M, Kautschitsch, S, Sommergruber, W, Tontsch-Grunt, U, Garin-Chesa, P, Bader, G, Zoephel, A, Quant, J, Heckel, A, Rettig, W.J. | | Deposit date: | 2008-02-08 | | Release date: | 2008-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | BIBF 1120: triple angiokinase inhibitor with sustained receptor blockade and good antitumor efficacy.
Cancer Res., 68, 2008
|
|
1JQH
 
 | | IGF-1 receptor kinase domain | | Descriptor: | IGF-1 receptor kinase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Pautsch, A, Zoephel, A, Ahorn, H, Spevak, W, Hauptmann, R, Nar, H. | | Deposit date: | 2001-08-07 | | Release date: | 2002-04-19 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of bisphosphorylated IGF-1 receptor kinase: insight into domain movements upon kinase activation.
Structure, 9, 2001
|
|
5K3Y
 
 | | Crystal structure of AuroraB/INCENP in complex with BI 811283 | | Descriptor: | Aurora kinase B-A, Inner centromere protein A, N-methyl-N-(1-methylpiperidin-4-yl)-4-{[4-({(1R,2S)-2-[(propan-2-yl)carbamoyl]cyclopentyl}amino)-5-(trifluoromethyl)pyrimidin-2-yl]amino}benzamide | | Authors: | Bader, G, Zahn, S.K, Zoephel, A. | | Deposit date: | 2016-05-20 | | Release date: | 2016-08-17 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pharmacological Profile of BI 847325, an Orally Bioavailable, ATP-Competitive Inhibitor of MEK and Aurora Kinases.
Mol.Cancer Ther., 15, 2016
|
|
6HAZ
 
 | | Crystal structure of the bromodomain of human SMARCA2 in complex with SMARCA-BD ligand | | Descriptor: | 2-(6-azanyl-5-piperazin-4-ium-1-yl-pyridazin-3-yl)phenol, Probable global transcription activator SNF2L2, ZINC ION | | Authors: | Bader, G, Steurer, S, Weiss-Puxbaum, A, Zoephel, A, Roy, M, Ciulli, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Nat.Chem.Biol., 15, 2019
|
|
5MW2
 
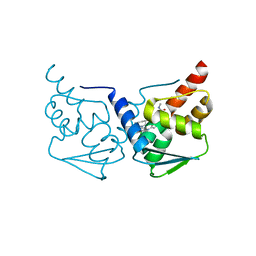 | | CRYSTAL STRUCTURE OF BCL-6 BTB-domain with BI-3802 | | Descriptor: | 2-[6-[[5-chloranyl-2-[(3~{S},5~{R})-3,5-dimethylpiperidin-1-yl]pyrimidin-4-yl]amino]-1-methyl-2-oxidanylidene-quinolin-3-yl]oxy-~{N}-methyl-ethanamide, B-cell lymphoma 6 protein | | Authors: | Bader, G, Flotzinger, G, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
6I8Z
 
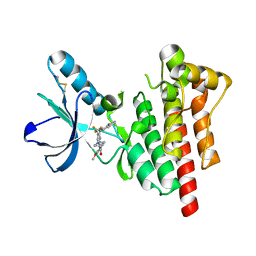 | | Crystal structure of PTK2 in complex with BI-4464. | | Descriptor: | 3-methoxy-~{N}-(1-methylpiperidin-1-ium-4-yl)-4-[[4-[(3-oxidanylidene-1,2-dihydroinden-4-yl)oxy]-5-(trifluoromethyl)pyrimidin-2-yl]amino]benzamide, Focal adhesion kinase 1 | | Authors: | Bader, G, Zoephel, A. | | Deposit date: | 2018-11-21 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Highly Selective PTK2 Proteolysis Targeting Chimeras to Probe Focal Adhesion Kinase Scaffolding Functions.
J.Med.Chem., 62, 2019
|
|
5MWD
 
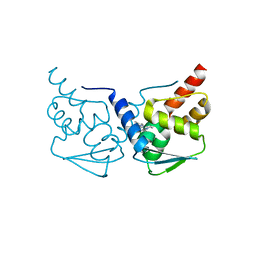 | | Crystal structure of the BCL6 BTB-domain with compound 2 | | Descriptor: | 5-[[5-chloranyl-2-(3,5-dimethylpyrazol-1-yl)pyrimidin-4-yl]amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein | | Authors: | Bader, G, Flotzinger, G, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
4G5J
 
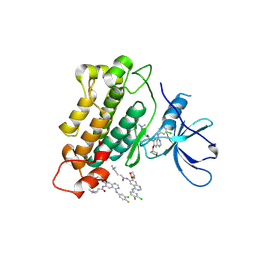 | | Crystal structure of EGFR kinase in complex with BIBW2992 | | Descriptor: | (2E)-N-{4-[(3-chloro-4-fluorophenyl)amino]-7-[(3S)-tetrahydrofuran-3-yloxy]quinazolin-6-yl}-4-(dimethylamino)but-2-enamide, Epidermal growth factor receptor, N-{4-[(3-chloro-4-fluorophenyl)amino]-7-[(3S)-tetrahydrofuran-3-yloxy]quinazolin-6-yl}-4-(dimethylamino)butanamide | | Authors: | Solca, F, Dahl, G, Zoephel, A, Bader, G, Sanderson, M, Klein, C, Kraemer, O, Himmelsbach, F, Haaksma, E, Adolf, G.R. | | Deposit date: | 2012-07-18 | | Release date: | 2012-08-29 | | Last modified: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Target Binding Properties and Cellular Activity of Afatinib (BIBW 2992), an Irreversible ErbB Family Blocker.
J.Pharmacol.Exp.Ther., 343, 2012
|
|
4G5P
 
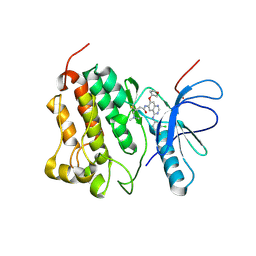 | | Crystal structure of EGFR kinase T790M in complex with BIBW2992 | | Descriptor: | Epidermal growth factor receptor, N-{4-[(3-chloro-4-fluorophenyl)amino]-7-[(3S)-tetrahydrofuran-3-yloxy]quinazolin-6-yl}-4-(dimethylamino)butanamide | | Authors: | Solca, F, Dahl, G, Zoephel, A, Bader, G, Sanderson, M, Klein, C, Kraemer, O, Himmelsbach, F, Haaksma, E, Adolf, G.R. | | Deposit date: | 2012-07-18 | | Release date: | 2012-08-29 | | Last modified: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Target Binding Properties and Cellular Activity of Afatinib (BIBW 2992), an Irreversible ErbB Family Blocker.
J.Pharmacol.Exp.Ther., 343, 2012
|
|
6RJ3
 
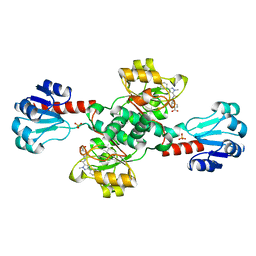 | | Crystal structure of PHGDH in complex with compound 15 | | Descriptor: | 4-[(1~{R})-1-[(2-methyl-5-phenyl-pyrazol-3-yl)carbonylamino]ethyl]benzoic acid, D-3-phosphoglycerate dehydrogenase, SULFATE ION | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
6RJ5
 
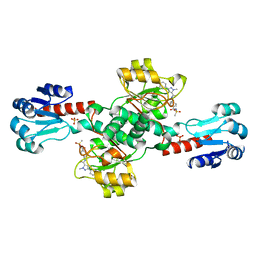 | | Crystal structure of PHGDH in complex with compound 39 | | Descriptor: | 2-methyl-~{N}-[(1~{R})-1-[4-(methylsulfonylcarbamoyl)phenyl]ethyl]-5-phenyl-pyrazole-3-carboxamide, D-3-phosphoglycerate dehydrogenase, SULFATE ION | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
6RIH
 
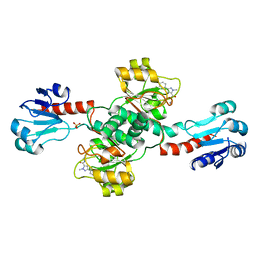 | | Crystal structure of PHGDH in complex with compound 9 | | Descriptor: | D-3-phosphoglycerate dehydrogenase, SULFATE ION, ~{N}-cyclopropyl-2-methyl-5-phenyl-pyrazole-3-carboxamide | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
6RJ2
 
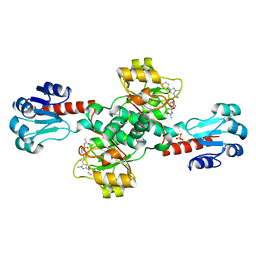 | | Crystal structure of PHGDH in complex with compound 40 | | Descriptor: | D-3-phosphoglycerate dehydrogenase, SULFATE ION, ~{N}-[(1~{R})-1-[4-(ethanoylsulfamoyl)phenyl]ethyl]-2-methyl-5-phenyl-pyrazole-3-carboxamide | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
6RJ6
 
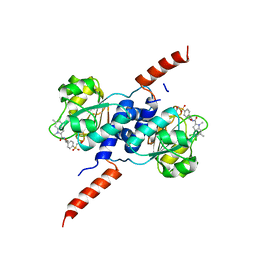 | | Crystal structure of PHGDH in complex with BI-4924 | | Descriptor: | 2-[4-[(1~{S})-1-[[4,5-bis(chloranyl)-1,6-dimethyl-indol-2-yl]carbonylamino]-2-oxidanyl-ethyl]phenyl]sulfonylethanoic acid, D-3-phosphoglycerate dehydrogenase | | Authors: | Bader, G, Wolkerstorfer, B, Zoephel, A. | | Deposit date: | 2019-04-26 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Intracellular Trapping of the Selective Phosphoglycerate Dehydrogenase (PHGDH) InhibitorBI-4924Disrupts Serine Biosynthesis.
J.Med.Chem., 62, 2019
|
|
5EU1
 
 | | CRYSTAL STRUCTURE OF BRD9 IN COMPLEX WITH BI-7273 | | Descriptor: | 4-[4-[(dimethylamino)methyl]-3,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, BRD9 | | Authors: | Bader, G, Martin, L.M, Steurer, S, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
5F25
 
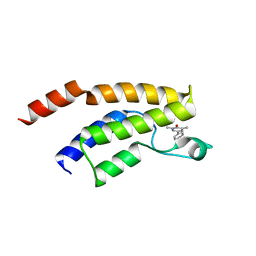 | | Crystal structure of the BRD9 bromodomain in complex with compound 4. | | Descriptor: | 4-(1,5-dimethyl-6-oxidanylidene-pyridin-3-yl)benzamide, BRD9 | | Authors: | Bader, G, Martin, L.J, Steurer, S, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2015-12-01 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
5F1H
 
 | | Crystal structure of the BRD9 bromodamian in complex with BI-9564. | | Descriptor: | 4-[4-[(dimethylamino)methyl]-2,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 9 | | Authors: | Bader, G, Martin, L.J, Steurer, S, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
5F1L
 
 | | Crystal structure of the bromodomain of BRD9 in complex with compound 9. | | Descriptor: | 5-[3,5-dimethoxy-4-[(3-oxidanylazetidin-1-yl)methyl]phenyl]-1,3-dimethyl-pyridin-2-one, Bromodomain-containing protein 9 | | Authors: | Bader, G, Martin, L.J, Steurer, S, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
5EYK
 
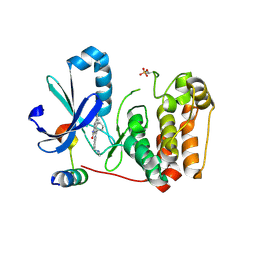 | | CRYSTAL STRUCTURE OF AURORA B IN COMPLEX WITH BI 847325 | | Descriptor: | 3-[(3~{Z})-3-[[[4-[(dimethylamino)methyl]phenyl]amino]-phenyl-methylidene]-2-oxidanylidene-1~{H}-indol-6-yl]-~{N}-ethyl-prop-2-ynamide, Aurora kinase B-A, Inner centromere protein A | | Authors: | Bader, G, Zoephel, A. | | Deposit date: | 2015-11-25 | | Release date: | 2016-08-17 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Pharmacological Profile of BI 847325, an Orally Bioavailable, ATP-Competitive Inhibitor of MEK and Aurora Kinases.
Mol.Cancer Ther., 15, 2016
|
|
5F2P
 
 | | Crystal structure of the BRD9 bromodomain in complex with compound 3. | | Descriptor: | 2-(dimethylamino)-6-methyl-pyrido[4,3-d]pyrimidin-5-one, BRD9 | | Authors: | Nar, H, Fiegen, D, Zoephel, A, Bader, G. | | Deposit date: | 2015-12-02 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
6G24
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound 3 | | Descriptor: | 2-[(~{E})-2-thiophen-2-ylethenyl]benzoic acid, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
6G2C
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound 9 | | Descriptor: | 3,5-dimethyl-4-(1-methyl-5-pyridin-4-yl-imidazol-4-yl)-1,2-oxazole, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
6G25
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound 4 | | Descriptor: | 3,5-dimethyl-4-(4-pyridin-4-yl-1~{H}-pyrazol-3-yl)-1,2-oxazole, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.432 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
6G2B
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound 8 | | Descriptor: | 4-(3-methyl-5-phenyl-imidazol-4-yl)pyridine, Histone-lysine N-methyltransferase NSD3 | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
