2ON7
 
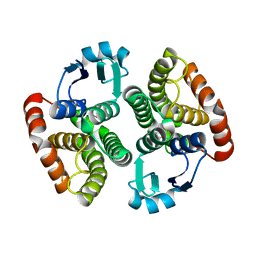 | | Structure of NaGST-1 | | Descriptor: | Na Glutathione S-transferase 1 | | Authors: | Asojo, O.A, Ngamelue, M, Homma, H, Goud, G, Zhan, B, Hotez, P.J. | | Deposit date: | 2007-01-23 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of Na-GST-1 and Na-GST-2 two glutathione s-transferase from the human hookworm Necator americanus
Bmc Struct.Biol., 7, 2007
|
|
2ON5
 
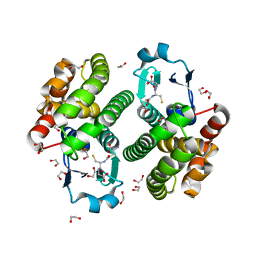 | | Structure of NaGST-2 | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Na Glutathione S-transferase 2 | | Authors: | Asojo, O.A, Ngamelue, M, Homma, H, Goud, G, Zhan, B, Hotez, P.J. | | Deposit date: | 2007-01-23 | | Release date: | 2007-08-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of Na-GST-1 and Na-GST-2 two glutathione s-transferase from the human hookworm Necator americanus
Bmc Struct.Biol., 7, 2007
|
|
7WNT
 
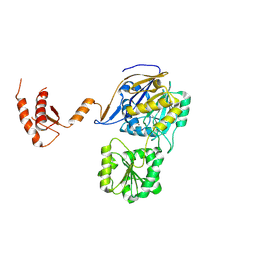 | | RNase J from Mycobacterium tuberculosis | | Descriptor: | Ribonuclease J, ZINC ION | | Authors: | Li, J, Bao, L, Hu, J, Zhan, B, Li, Z. | | Deposit date: | 2022-01-19 | | Release date: | 2023-01-25 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural insights into RNase J that plays an essential role in Mycobacterium tuberculosis RNA metabolism
Nat Commun, 14, 2023
|
|
7WNU
 
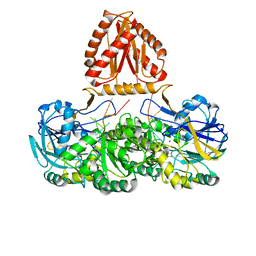 | | Mycobacterium tuberculosis Rnase J complex with 7nt RNA | | Descriptor: | RNA (5'-R(P*AP*AP*AP*AP*AP*AP*A)-3'), Ribonuclease J, ZINC ION | | Authors: | Li, J, Hu, J, Bao, L, Zhan, B, Li, Z. | | Deposit date: | 2022-01-19 | | Release date: | 2023-01-25 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into RNase J that plays an essential role in Mycobacterium tuberculosis RNA metabolism
Nat Commun, 14, 2023
|
|
6PWV
 
 | | Cryo-EM structure of MLL1 core complex bound to the nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
4M58
 
 | | Crystal Structure of an transition metal transporter | | Descriptor: | Cobalamin biosynthesis protein CbiM, NICKEL (II) ION | | Authors: | Yu, Y, Yan, C.Y, Zhang, B, Li, X.L, Gu, J.K. | | Deposit date: | 2013-08-08 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Planar substrate-binding site dictates the specificity of ECF-type nickel/cobalt transporters
Cell Res., 24, 2014
|
|
5XMY
 
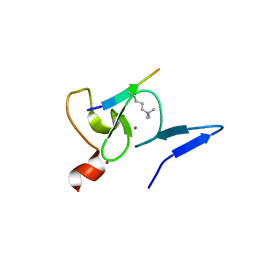 | |
6PWW
 
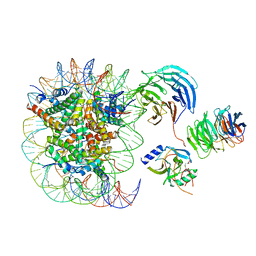 | | Cryo-EM structure of MLL1 in complex with RbBP5 and WDR5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
6PWX
 
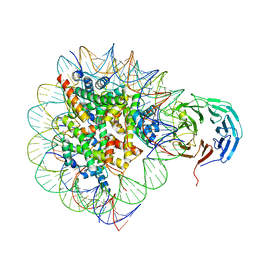 | | Cryo-EM structure of RbBP5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
4UUQ
 
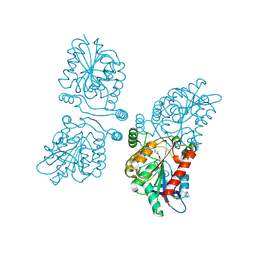 | | Crystal structure of human mono-glyceride lipase in complex with SAR127303 | | Descriptor: | 4-({[(4-chlorophenyl)sulfonyl]amino}methyl)piperidine-1-carboxylic acid, MONOGLYCERIDE LIPASE | | Authors: | Griebel, G, Pichat, P, Beeske, S, Leroy, T, Redon, N, Francon, D, Bert, L, Even, L, Lopez-Grancha, M, Tolstykh, T, Sun, F, Yu, Q, Brittain, S, Arlt, H, He, T, Zhang, B, Wiederschain, D, Bertrand, T, Houtman, J, Rak, A, Vallee, F, Michot, N, Auge, F, Menet, V, Bergis, O.E, George, P, Avenet, P, Mikol, V, Didier, M, Escoubet, J. | | Deposit date: | 2014-07-30 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Selective Blockade of the Hydrolysis of the Endocannabinoid 2-Arachidonoylglycerol Impairs Learning and Memory Performance While Producing Antinociceptive Activity in Rodents.
Sci.Rep., 5, 2015
|
|
7BUY
 
 | | The crystal structure of COVID-19 main protease in complex with carmofur | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, hexylcarbamic acid | | Authors: | Zhao, Y, Zhang, B, Jin, Z, Liu, X, Yang, H, Rao, Z. | | Deposit date: | 2020-04-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7BQY
 
 | | THE CRYSTAL STRUCTURE OF COVID-19 MAIN PROTEASE IN COMPLEX WITH AN INHIBITOR N3 at 1.7 angstrom | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
6PFM
 
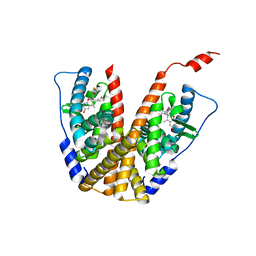 | | Crystal structure of GDC-0927 bound to estrogen receptor alpha | | Descriptor: | (2S)-2-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-3-(3-hydroxyphenyl)-4-methyl-2H-1-benzopyran-6-ol, Estrogen receptor | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Wang, X, Zbieg, J.R, Labadie, S.S, Li, J, Ray, N.C, Ortwine, D. | | Deposit date: | 2019-06-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of a C-8 hydroxychromene as a potent degrader of estrogen receptor alpha with improved rat oral exposure over GDC-0927.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6XVH
 
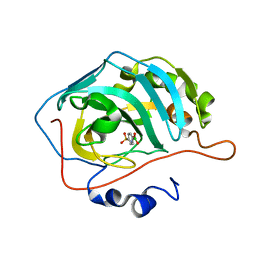 | | carbonic anhydrase with bound arylsulfonamide- boronic acid | | Descriptor: | (4-sulfamoylphenyl)boronic acid, Carbonic anhydrase 2, ZINC ION | | Authors: | Perez, C, Zhang, B. | | Deposit date: | 2020-01-22 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An easy to assemble, fully modulable, selectiv and
colorimetric artificial bio-sensor for fingerprintting
the recognition of neurotransmitters
To Be Published
|
|
6CB6
 
 | |
6CB7
 
 | |
6LU7
 
 | | The crystal structure of COVID-19 main protease in complex with an inhibitor N3 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | Deposit date: | 2020-01-26 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
5MK5
 
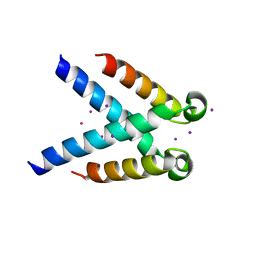 | | Structures of DHBN domain of human BLM helicase | | Descriptor: | Bloom syndrome protein, IODIDE ION, POTASSIUM ION | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-12-02 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
7ZHI
 
 | |
7ZGS
 
 | |
7ZP2
 
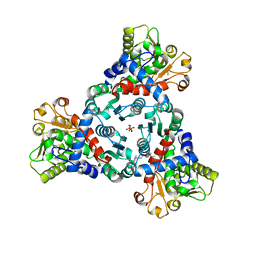 | | Crystal Structure of truncated aspartate transcarbamoylase from Plasmodium falciparum in complex with BDA-04 | | Descriptor: | Aspartate carbamoyltransferase, SODIUM ION, SULFATE ION, ... | | Authors: | Wang, C, Zhang, B, Groves, M.R. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Discovery of Small-Molecule Allosteric Inhibitors of Pf ATC as Antimalarials.
J.Am.Chem.Soc., 144, 2022
|
|
7ZST
 
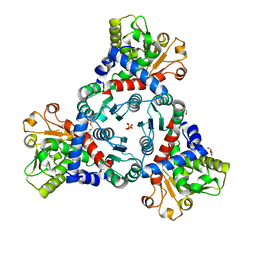 | | Crystal Structure of truncated aspartate transcarbamoylase from Plasmodium falciparum in complex with FLA-01 | | Descriptor: | 2-azanyl-~{N}-(2-methoxyethyl)-5-phenyl-thiophene-3-carboxamide, Aspartate carbamoyltransferase, GLYCEROL, ... | | Authors: | Wang, C, Zhang, B, Groves, M.R. | | Deposit date: | 2022-05-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Small-Molecule Allosteric Inhibitors of Pf ATC as Antimalarials.
J.Am.Chem.Soc., 144, 2022
|
|
7ZID
 
 | |
4S1R
 
 | | Crystal structure of a VRC01-lineage antibody, 45-VRC01.H08.F-117225, in complex with clade A/E HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab of VRC01 light chain, Fab of VRC01-lineage antibody,45-VRC01.H08.F-117225 heavy chain, ... | | Authors: | Kwon, Y.D, Yang, Y, Zhang, B, Kwong, P.D. | | Deposit date: | 2015-01-14 | | Release date: | 2015-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.214 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell(Cambridge,Mass.), 161, 2015
|
|
4S1Q
 
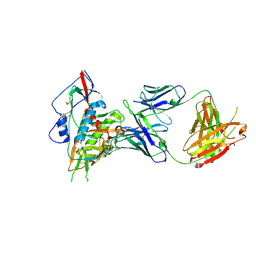 | | Crystal structure of a VRC01-lineage antibody, 45-VRC01.H03+06.D-001739, in complex with clade A/E HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab of VRC01 light chain, Fab of VRC01-lineage antibody,45-VRC01.H03+06.D-001739 heavy chain, ... | | Authors: | Kwon, Y.D, Yang, Y, Zhang, B, Kwong, P.D. | | Deposit date: | 2015-01-14 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection.
Cell(Cambridge,Mass.), 161, 2015
|
|
