1ZYZ
 
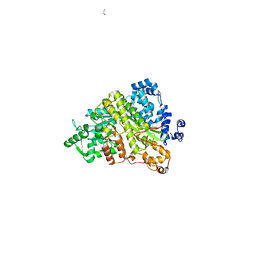 | | Structures of Yeast Ribonucloetide Reductase I | | 分子名称: | GLYCINE, Ribonucleoside-diphosphate reductase large chain 1 | | 著者 | Xu, H, Faber, C, Uchiki, T, Fairman, J.W, Racca, J, Dealwis, C. | | 登録日 | 2005-06-13 | | 公開日 | 2006-03-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1ZZD
 
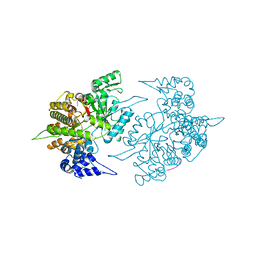 | | Structures of Yeast Ribonucleotide Reductase I | | 分子名称: | Ribonucleoside-diphosphate reductase large chain 1, Ribonucleoside-diphosphate reductase small chain 2 | | 著者 | Xu, H, Faber, C, Uchiki, T, Fairman, J.W, Racca, J, Dealwis, C. | | 登録日 | 2005-06-13 | | 公開日 | 2006-03-07 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GZY
 
 | |
2GZZ
 
 | |
6LG7
 
 | |
6LG6
 
 | |
6N4Q
 
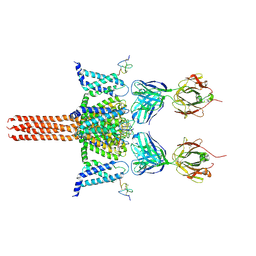 | | CryoEM structure of Nav1.7 VSD2 (actived state) in complex with the gating modifier toxin ProTx2 | | 分子名称: | Beta/omega-theraphotoxin-Tp2a, Fab heavy chain, Fab light chain, ... | | 著者 | Xu, H, Rohou, A, Arthur, C.P, Estevez, A, Ciferri, C, Payandeh, J, Koth, C.M. | | 登録日 | 2018-11-20 | | 公開日 | 2019-01-23 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin.
Cell, 176, 2019
|
|
6N4I
 
 | |
6N4R
 
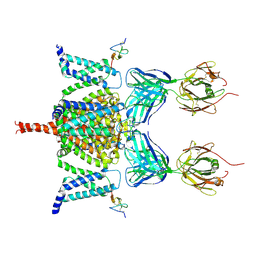 | | CryoEM structure of Nav1.7 VSD2 (deactived state) in complex with the gating modifier toxin ProTx2 | | 分子名称: | Beta/omega-theraphotoxin-Tp2a, Fab heavy chain, Fab light chain, ... | | 著者 | Xu, H, Rohou, A, Arthur, C.P, Estevez, A, Ciferri, C, Payandeh, J, Koth, C.M. | | 登録日 | 2018-11-20 | | 公開日 | 2019-01-23 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin.
Cell, 176, 2019
|
|
7DCN
 
 | |
7DCM
 
 | | Crystal structure of CITX | | 分子名称: | Probable apo-citrate lyase phosphoribosyl-dephospho-CoA transferase, ZINC ION | | 著者 | Xu, H, Wang, B, Su, X.D. | | 登録日 | 2020-10-26 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-05-17 | | 実験手法 | X-RAY DIFFRACTION (2.495 Å) | | 主引用文献 | Co-evolution-based prediction of metal-binding sites in proteomes by machine learning.
Nat.Chem.Biol., 19, 2023
|
|
7XHE
 
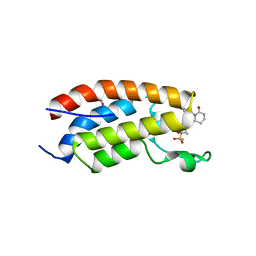 | | Crystal structure of CBP bromodomain liganded with CCS151 | | 分子名称: | (6S)-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(3R)-1-methylsulfonylpyrrolidin-3-yl]benzimidazol-2-yl]-1-(3-fluoranyl-4-methoxy-phenyl)piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | 著者 | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-04-08 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7XH6
 
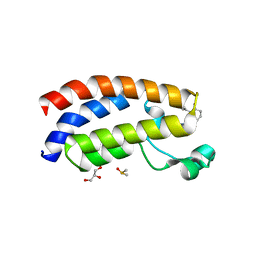 | | Crystal structure of CBP bromodomain liganded with CCS1477 | | 分子名称: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | 著者 | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-04-07 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7XI0
 
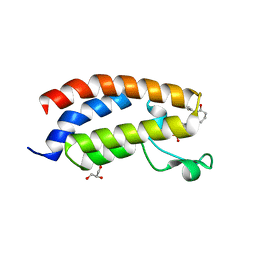 | | Crystal structure of CBP bromodomain liganded with CCS150 | | 分子名称: | (6S)-1-(3-chloranyl-4-methoxy-phenyl)-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(3R)-1-methylsulfonylpyrrolidin-3-yl]benzimidazol-2-yl]piperidin-2-one, CREB-binding protein, GLYCEROL | | 著者 | Xu, H, Xiang, Q, Wang, C, Zhang, C, Luo, G, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-04-11 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Structural insights revealed by the cocrystal structure of CCS1477 in complex with CBP bromodomain
Biochem.Biophys.Res.Commun., 623, 2022
|
|
7EK0
 
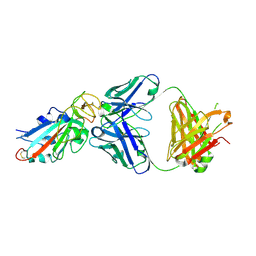 | | Complex Structure of antibody BD-503 and RBD-N501Y of COVID-19 | | 分子名称: | Heavy Chain of BD-503, Light Chain of BD-503, Spike protein S1 | | 著者 | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | 登録日 | 2021-04-03 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7EJZ
 
 | | Complex Structure of antibody BD-503 and RBD-S477N of COVID-19 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | 著者 | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | 登録日 | 2021-04-03 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.63 Å) | | 主引用文献 | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7EJY
 
 | | Complex Structure of antibody BD-503 and RBD of COVID-19 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | 著者 | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | 登録日 | 2021-04-03 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.04 Å) | | 主引用文献 | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7F6Z
 
 | | Complex Structure of antibody BD-503 and RBD-501Y.V2 of COVID-19 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | 著者 | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | 登録日 | 2021-06-26 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
7F6Y
 
 | | Complex Structure of antibody BD-503 and RBD-E484K of COVID-19 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of BD-503, Light Chain of BD-503, ... | | 著者 | Xu, H, Wang, B, Zhao, T.N, Su, X.D. | | 登録日 | 2021-06-26 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure-based analyses of neutralization antibodies interacting with naturally occurring SARS-CoV-2 RBD variants.
Cell Res., 31, 2021
|
|
4ORD
 
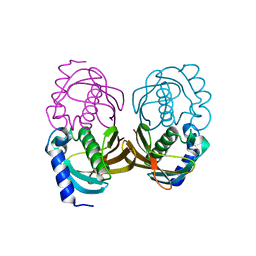 | |
2KRL
 
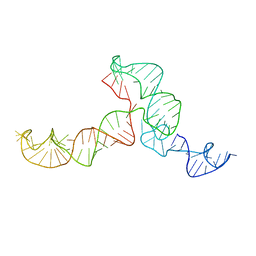 | | The ensemble of the solution global structures of the 102-nt ribosome binding structure element of the turnip crinkle virus 3' UTR RNA | | 分子名称: | RNA (102-MER) | | 著者 | Zuo, X, Wang, J, Yu, P, Eyler, D, Xu, H, Starich, M, Tiede, D, Simon, A, Kasprzak, W, Schwieters, C, Shapiro, B. | | 登録日 | 2009-12-18 | | 公開日 | 2011-02-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3' UTR of turnip crinkle virus.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7YL2
 
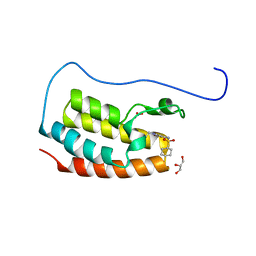 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004 | | 分子名称: | Bromodomain-containing protein 4, GLYCEROL, N-(1-ethyl-2-oxidanylidene-3H-indol-5-yl)cyclohexanesulfonamide, ... | | 著者 | Huang, Y, Wei, A, Dong, R, Xu, H, Zhang, C, Chen, Z, Li, J, Wu, X, Zhang, Y, Xu, Y. | | 登録日 | 2022-07-25 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor Y07004
To Be Published
|
|
3BZU
 
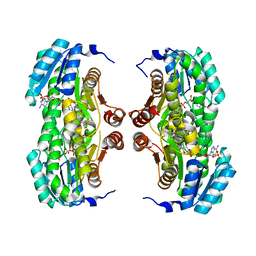 | | Crystal structure of human 11-beta-hydroxysteroid dehydrogenase(HSD1) in complex with NADP and thiazolone inhibitor | | 分子名称: | (5S)-2-{[(1S)-1-(2-fluorophenyl)ethyl]amino}-5-methyl-5-(trifluoromethyl)-1,3-thiazol-4(5H)-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Min, X, Sudom, A, Xu, H, Wang, Z, Walker, N.P. | | 登録日 | 2008-01-18 | | 公開日 | 2008-06-10 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural characterization and pharmacodynamic effects of an orally active 11beta-hydroxysteroid dehydrogenase type 1 inhibitor.
Chem.Biol.Drug Des., 71, 2008
|
|
5GW8
 
 | | Crystal structure of a putative DAG-like lipase (MgMDL2) from Malassezia globosa | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | 著者 | Xu, J, Xu, H, Liu, J. | | 登録日 | 2016-09-09 | | 公開日 | 2017-09-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Malassezia globosa MgMDL2 lipase: Crystal structure and rational modification of substrate specificity.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
7YHN
 
 | | ANTI-TUMOR AGENT Y48 IN COMPLEX WITH TUBULIN | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-methyl-3-[(4-methylphenyl)sulfonylamino]-~{N}-[(6-methylpyridin-3-yl)methyl]benzamide, GUANOSINE-5'-DIPHOSPHATE, ... | | 著者 | Du, T, Ji, M, Hou, Z, Lin, S, Zhang, J, Wu, D, Zhang, K, Lu, D, Xu, H, Chen, X. | | 登録日 | 2022-07-14 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Optimization of Benzamide Derivatives as Potent and Orally Active Tubulin Inhibitors Targeting the Colchicine Binding Site.
J.Med.Chem., 65, 2022
|
|
