5HOI
 
 | |
5HOG
 
 | |
6MDR
 
 | | Cryo-EM structure of the Ceru+32/GFP-17 protomer | | Descriptor: | Ceru+32, GFP-17 | | Authors: | Simon, A.J, Zhou, Y, Ramasubramani, V, Glaser, J, Pothukuchy, A, Golihar, J, Gerberich, J.C, Leggere, J.C, Morrow, B.R, Jung, C, Glotzer, S.C, Taylor, D.W, Ellington, A.D. | | Deposit date: | 2018-09-05 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Supercharging enables organized assembly of synthetic biomolecules.
Nat Chem, 11, 2019
|
|
5DGO
 
 | |
4C93
 
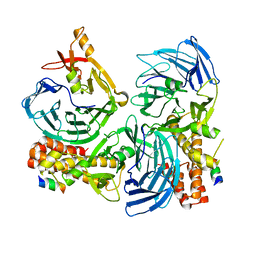 | |
4C8H
 
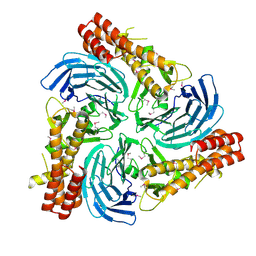 | |
4C95
 
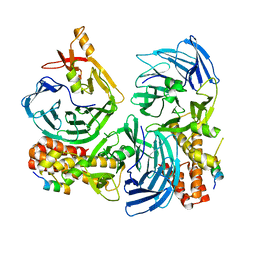 | |
4C8S
 
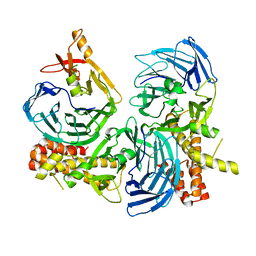 | |
4A20
 
 | | Crystal structure of the Ubl domain of Mdy2 (Get5) at 1.78A | | Descriptor: | SULFATE ION, UBIQUITIN-LIKE PROTEIN MDY2 | | Authors: | Simon, A.C, Simpson, P.J, Murray, J.W, Isaacson, R.L. | | Deposit date: | 2011-09-21 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of the Sgt2/Get5 Complex Provides Insights Into Get-Mediated Targeting of Tail-Anchored Membrane Proteins
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4ASW
 
 | | Structure of the complex between the N-terminal dimerisation domain of Sgt2 and the UBL domain of Get5 | | Descriptor: | SMALL GLUTAMINE-RICH TETRATRICOPEPTIDE REPEAT-CONTAINING PROTEIN 2, UBIQUITIN-LIKE PROTEIN MDY2 | | Authors: | Simon, A.C, Simpson, P.J, Goldstone, R.M, Krysztofinska, E.M, Murray, J.W, High, S, Isaacson, R.L. | | Deposit date: | 2012-05-03 | | Release date: | 2013-01-16 | | Last modified: | 2021-06-23 | | Method: | SOLUTION NMR | | Cite: | Structure of the Sgt2/Get5 Complex Provides Insights Into Get-Mediated Targeting of Tail-Anchored Membrane Proteins
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4ASV
 
 | | Solution structure of the N-terminal dimerisation domain of Sgt2 | | Descriptor: | SMALL GLUTAMINE-RICH TETRATRICOPEPTIDE REPEAT-CONTAINING PROTEIN 2 | | Authors: | Simon, A.C, Simpson, P.J, Goldstone, R.M, Krysztofinska, E.M, Murray, J.W, High, S, Isaacson, R.L. | | Deposit date: | 2012-05-03 | | Release date: | 2013-01-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the Sgt2/Get5 Complex Provides Insights Into Get-Mediated Targeting of Tail-Anchored Membrane Proteins
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8ADT
 
 | | Rational design of a calcium-independent trypsin variant | | Descriptor: | Serine protease 1 | | Authors: | Simon, A.H, Liebscher, S, Kattner, A, Kattner, C, Bordusa, F. | | Deposit date: | 2022-07-11 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rational Design of a Calcium-Independent Trypsin Variant
Catalysts, 12, 2022
|
|
3Q3Z
 
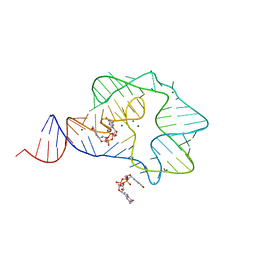 | | Structure of a c-di-GMP-II riboswitch from C. acetobutylicum bound to c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, c-di-GMP-II riboswitch | | Authors: | Smith, K.D, Shanahan, C.A, Moore, E.L, Simon, A.C, Strobel, S.A. | | Deposit date: | 2010-12-22 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis of differential ligand recognition by two classes of bis-(3'-5')-cyclic dimeric guanosine monophosphate-binding riboswitches.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6HNJ
 
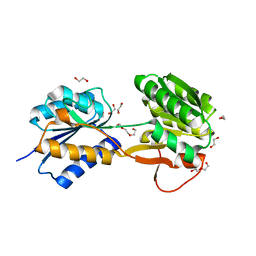 | | The ligand-bound, open structure of CD0873, a substrate binding protein with adhesive properties from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, ABC-type transport system, sugar-family extracellular solute-binding protein, ... | | Authors: | Bradshaw, W.J, Kovacs-Simon, A, Harmer, N.J, Michell, S.L, Acharya, K.R. | | Deposit date: | 2018-09-15 | | Release date: | 2019-08-28 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular features of lipoprotein CD0873: A potential vaccine against the human pathogenClostridioides difficile.
J.Biol.Chem., 294, 2019
|
|
3S64
 
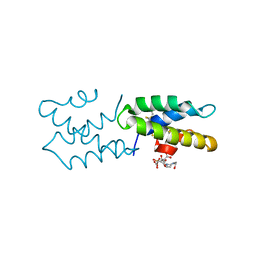 | | Saposin-like protein Ac-SLP-1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CITRIC ACID, Saposin-like protein 1 | | Authors: | Willis, C, Wang, C.K, Osman, A, Simon, A, Mulvenna, J, Pickering, D, Riboldi-Tunicliffe, A, Jones, M.K, Loukas, A, Hofmann, A. | | Deposit date: | 2011-05-25 | | Release date: | 2012-01-18 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Insights into the membrane interactions of the saposin-like proteins Na-SLP-1 and Ac-SLP-1 from human and dog hookworm.
Plos One, 6, 2011
|
|
3S63
 
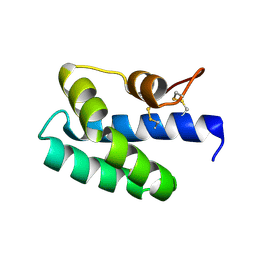 | | Saposin-like protein Na-SLP-1 | | Descriptor: | Saposin-like protein | | Authors: | Willis, C, Wang, C.K, Osman, A, Simon, A, Mulvenna, J, Pickering, D, Riboldi-Tunicliffe, A, Jones, M.K, Loukas, A, Hofmann, A. | | Deposit date: | 2011-05-24 | | Release date: | 2012-01-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the membrane interactions of the saposin-like proteins Na-SLP-1 and Ac-SLP-1 from human and dog hookworm.
Plos One, 6, 2011
|
|
3ZPK
 
 | | Atomic-resolution structure of a quadruplet cross-beta amyloid fibril. | | Descriptor: | TRANSTHYRETIN | | Authors: | Fitzpatrick, A.W.P, Debelouchina, G.T, Bayro, M.J, Clare, D.K, Caporini, M.A, Bajaj, V.S, Jaroniec, C.P, Wang, L, Ladizhansky, V, Muller, S.A, MacPhee, C.E, Waudby, C.A, Mott, H.R, de Simone, A, Knowles, T.P.J, Saibil, H.R, Vendruscolo, M, Orlova, E.V, Griffin, R.G, Dobson, C.M. | | Deposit date: | 2013-02-28 | | Release date: | 2013-12-04 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY, SOLID-STATE NMR | | Cite: | Atomic Structure and Hierarchical Assembly of a Cross-Beta Amyloid Fibril.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6HNI
 
 | | The ligand-bound, closed structure of CD0873, a substrate binding protein with adhesive properties from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, ABC-type transport system, sugar-family extracellular solute-binding protein, ... | | Authors: | Bradshaw, W.J, Kovacs-Simon, A, Harmer, N.J, Michell, S.L, Acharya, K.R. | | Deposit date: | 2018-09-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Molecular features of lipoprotein CD0873: A potential vaccine against the human pathogenClostridioides difficile.
J.Biol.Chem., 294, 2019
|
|
8RI9
 
 | | Late alpha-Synuclein fibril structure from liquid-liquid phase separations. | | Descriptor: | Alpha-synuclein | | Authors: | De Simone, A, Barritt, J.D, Chen, S, Cascella, R, Cecchi, C, Bigi, A, Jarvis, J.A, Chiti, F, Dobson, C.M, Fusco, G. | | Deposit date: | 2023-12-18 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-Toxicity Relationship in Intermediate Fibrils from alpha-Synuclein Condensates.
J.Am.Chem.Soc., 2024
|
|
5OGS
 
 | | Crystal structure of human AND-1 SepB domain | | Descriptor: | MALONATE ION, WD repeat and HMG-box DNA-binding protein 1 | | Authors: | Pellegrini, L, Simon, A.C. | | Deposit date: | 2017-07-13 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | The human CTF4-orthologue AND-1 interacts with DNA polymerase alpha /primase via its unique C-terminal HMG box.
Open Biol, 7, 2017
|
|
5E27
 
 | | The structure of Resuscitation Promoting Factor B from M. tuberculosis reveals unexpected ubiquitin-like domains | | Descriptor: | Resuscitation-promoting factor RpfB | | Authors: | Ruggiero, A, Squeglia, F, Romano, M, Vitagliano, L, De Simone, A, Berisio, R. | | Deposit date: | 2015-09-30 | | Release date: | 2015-11-18 | | Last modified: | 2015-12-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of Resuscitation promoting factor B from M. tuberculosis reveals unexpected ubiquitin-like domains.
Biochim.Biophys.Acta, 1860, 2015
|
|
2KRL
 
 | | The ensemble of the solution global structures of the 102-nt ribosome binding structure element of the turnip crinkle virus 3' UTR RNA | | Descriptor: | RNA (102-MER) | | Authors: | Zuo, X, Wang, J, Yu, P, Eyler, D, Xu, H, Starich, M, Tiede, D, Simon, A, Kasprzak, W, Schwieters, C, Shapiro, B. | | Deposit date: | 2009-12-18 | | Release date: | 2011-02-09 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3' UTR of turnip crinkle virus.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4EMN
 
 | | Crystal structure of RpfB catalytic domain in complex with benzamidine | | Descriptor: | BENZAMIDINE, Probable resuscitation-promoting factor rpfB, SULFATE ION | | Authors: | Ruggiero, A, Marchant, J, Squeglia, F, Makarov, V, De Simone, A, Berisio, R. | | Deposit date: | 2012-04-12 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Molecular determinants of inactivation of the resuscitation promoting factor B from Mycobacterium tuberculosis.
J.Biomol.Struct.Dyn., 31, 2013
|
|
6GJJ
 
 | | Cyclophilin A complexed with tri-vector ligand 2. | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, ethyl 2-[[(4-aminophenyl)methyl-propyl-carbamoyl]amino]ethanoate | | Authors: | Georgiou, C, De Simone, A, Walkinshaw, M.D, Michel, J. | | Deposit date: | 2018-05-16 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | A computationally designed binding mode flip leads to a novel class of potent tri-vector cyclophilin inhibitors.
Chem Sci, 10, 2019
|
|
6GJI
 
 | | Cyclophilin A complexed with the tri-vector ligand 8. | | Descriptor: | GLYCEROL, Peptidyl-prolyl cis-trans isomerase A, ethyl 2-[[(4-aminophenyl)methyl-[(1-methyl-1,2,3-triazol-4-yl)methyl]carbamoyl]amino]ethanoate | | Authors: | Georgiou, C, De Simone, A, Walkinshaw, M.D, Michel, J. | | Deposit date: | 2018-05-16 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A computationally designed binding mode flip leads to a novel class of potent tri-vector cyclophilin inhibitors.
Chem Sci, 10, 2019
|
|
