7CWA
 
 | | Crystal structure of PDE8A catalytic domain in complex with clofarabine | | Descriptor: | 2-CHLORO-9-(2-DEOXY-2-FLUORO-B -D-ARABINOFURANOSYL)-9H-PURIN-6-AMINE, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Huang, Y, Wu, X.-N, Zhou, Q, Wu, Y, Luo, H.-B. | | Deposit date: | 2020-08-27 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rational Design of 2-Chloroadenine Derivatives as Highly Selective Phosphodiesterase 8A Inhibitors.
J.Med.Chem., 63, 2020
|
|
3HI1
 
 | | Structure of HIV-1 gp120 (core with V3) in Complex with CD4-Binding-Site Antibody F105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F105 Heavy Chain, F105 Light Chain, ... | | Authors: | Kwon, Y.D, Chen, L, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z, Zhang, M.-Y, Arthos, J, Burton, D.R, Dimitrov, D, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-05-18 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
8H40
 
 | | Cryo-EM structure of the transcription activation complex NtcA-TAC | | Descriptor: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8H3V
 
 | | Cryo-EM structure of the full transcription activation complex NtcA-NtcB-TAC | | Descriptor: | DNA (125-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Han, S.J, Jiang, Y.L, You, L.L, Shen, L.Q, Wu, X.X, Yang, F, Kong, W.W, Chen, Z.P, Zhang, Y, Zhou, C.Z. | | Deposit date: | 2022-10-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | DNA looping mediates cooperative transcription activation.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3IDX
 
 | | Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C222 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fab b13 heavy chain, ... | | Authors: | Chen, L, Kwon, Y.D, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z.Y, Zhang, M.Y, Arthos, J, Burton, D.R, Dimitrov, D.S, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
3IDY
 
 | | Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C2221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab b13 heavy chain, Fab b13 light chain, ... | | Authors: | Chen, L, Kwon, Y.D, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z.Y, Zhang, M.Y, Arthos, J, Burton, D.R, Dimitrov, D.S, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
2IC1
 
 | | Crystal Structure of Human Cysteine Dioxygenase in Complex with Substrate Cysteine | | Descriptor: | CYSTEINE, Cysteine dioxygenase type 1, FE (II) ION | | Authors: | Ye, S, Wu, X, Wei, L, Tang, D, Sun, P, Rao, Z. | | Deposit date: | 2006-09-12 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An Insight into the Mechanism of Human Cysteine Dioxygenase: KEY ROLES OF THE THIOETHER-BONDED TYROSINE-CYSTEINE COFACTOR.
J.Biol.Chem., 282, 2007
|
|
3R0M
 
 | | Crystal structure of anti-HIV llama VHH antibody A12 | | Descriptor: | Llama VHH A12, SULFATE ION | | Authors: | Chen, L, McLellan, J.S, Kwon, Y.D, Schmidt, S, Wu, X, Zhou, T, Yang, Y, Zhang, B, Forsman, A, Weiss, R.A, Verrips, T, Mascola, J, Kwong, P.D. | | Deposit date: | 2011-03-08 | | Release date: | 2012-03-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Single-Headed Immunoglobulins Efficiently Penetrate CD4-Binding Site and Effectively Neutralize HIV-1
To be Published
|
|
2JFD
 
 | | Structure of the MAT domain of human FAS | | Descriptor: | FATTY ACID SYNTHASE | | Authors: | Bunkoczi, G, Kavanagh, K, Hozjan, V, Rojkova, A, Wu, X, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Smith, S, Oppermann, U. | | Deposit date: | 2007-01-31 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for different specificities of acyltransferases associated with the human cytosolic and mitochondrial fatty acid synthases.
Chem. Biol., 16, 2009
|
|
3RJQ
 
 | | Crystal structure of anti-HIV llama VHH antibody A12 in complex with C186 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C186 gp120, Llama VHH A12 | | Authors: | Chen, L, McLellan, J.S, Kwon, Y.D, Schmidt, S, Wu, X, Zhou, T, Yang, Y, Zhang, B, Forsman, A, Weiss, R.A, Verrips, T, Mascola, J, Kwong, P.D. | | Deposit date: | 2011-04-15 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Crystal structure of anti-HIV A12 VHH of llama antibody in complex with C1086 gp120
To be Published
|
|
2JFK
 
 | | Structure of the MAT domain of human FAS with malonyl-CoA | | Descriptor: | COENZYME A, FATTY ACID SYNTHASE | | Authors: | Bunkoczi, G, Kavanagh, K, Hozjan, V, Rojkova, A, Watt, S, Wu, X, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Smith, S, Oppermann, U. | | Deposit date: | 2007-02-02 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the MAT Domain of Human Fas with Malonyl-Coa
To be Published
|
|
6J1L
 
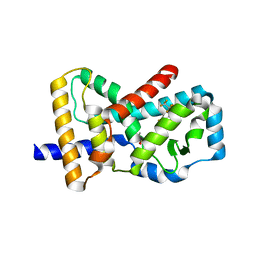 | | Crystal Structure Analysis of the ROR gamma(C455E) | | Descriptor: | 2-[4-(ethylsulfonyl)phenyl]-N-[2'-fluoro-4'-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)[1,1'-biphenyl]-4-yl]acetamide, Nuclear receptor ROR-gamma | | Authors: | zhang, Y, Li, C.C, wu, X.S. | | Deposit date: | 2018-12-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Characterization of XY101, a Potent, Selective, and Orally Bioavailable ROR gamma Inverse Agonist for Treatment of Castration-Resistant Prostate Cancer.
J.Med.Chem., 62, 2019
|
|
4ZUZ
 
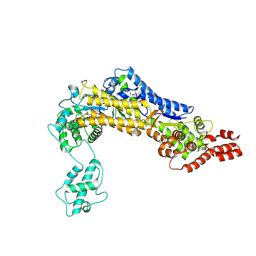 | | SidC 1-871 | | Descriptor: | SidC | | Authors: | Luo, X, Wasilko, D.J, Liu, Y, Sun, J, Wu, X, Luo, Z.-Q, Mao, Y. | | Deposit date: | 2015-05-18 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of the Legionella Virulence Factor, SidC Reveals a Unique PI(4)P-Specific Binding Domain Essential for Its Targeting to the Bacterial Phagosome.
Plos Pathog., 11, 2015
|
|
4FZE
 
 | |
7WHW
 
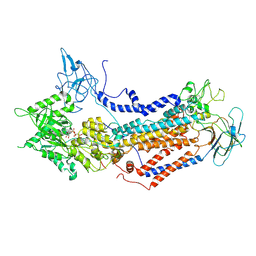 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in detergent with AMPPCP (E1-ATP state) | | Descriptor: | Alkylphosphocholine resistance protein LEM3, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7WHV
 
 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in detergent with beryllium fluoride (E2P state) | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Alkylphosphocholine resistance protein LEM3, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2021-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7XNE
 
 | | Crystal structure of CBP bromodomain liganded with Y08284 | | Descriptor: | CREB-binding protein, GLYCEROL, N-[3-(1-cyclopropylpyrazol-4-yl)-2-fluoranyl-5-[(1S)-1-oxidanylethyl]phenyl]-3-ethanoyl-7-methoxy-indolizine-1-carboxamide | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, C, Hu, Q, Luo, G, Hu, J, Zhuang, X, Zou, L, Shen, H, Wu, X, Zhang, Y, Kong, X, Xu, Y. | | Deposit date: | 2022-04-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
4HBN
 
 | | Crystal structure of the human HCN4 channel C-terminus carrying the S672R mutation | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, PHOSPHATE ION, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Xu, X, Marni, F, Wu, X, Su, Z, Musayev, F, Shrestha, S, Xie, C, Gao, W, Liu, Q, Zhou, L. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Local and Global Interpretations of a Disease-Causing Mutation near the Ligand Entry Path in Hyperpolarization-Activated cAMP-Gated Channel.
Structure, 20, 2012
|
|
4DKQ
 
 | | Crystal structure of clade A/E 93TH057 HIV-1 gp120 core in complex with DMJ-I-228 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, N-[(1S,2S)-2-carbamimidamido-2,3-dihydro-1H-inden-1-yl]-N'-(4-chloro-3-fluorophenyl)ethanediamide, ... | | Authors: | Kwon, Y.D, LaLonde, J.M, Jones, D.M, Sun, A.W, Courter, J.R, Soeta, T, Kobayashi, T, Princiotto, A.M, Wu, X, Mascola, J, Schon, A, Freire, E, Sodroski, J, Madani, N, Smith III, A.B, Kwong, P.D. | | Deposit date: | 2012-02-03 | | Release date: | 2012-05-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.888 Å) | | Cite: | Structure-Based Design, Synthesis, and Characterization of Dual Hotspot Small-Molecule HIV-1 Entry Inhibitors.
J.Med.Chem., 55, 2012
|
|
7EVJ
 
 | | Crystal structure of CBP bromodomain liganded with 9c | | Descriptor: | 3-acetyl-1-((3-(1-cyclopropyl-1H-pyrazol-4-yl)-2-fluoro-5-(hydroxymethyl)phenyl)carbamoyl)indolizin-7-yl dimethylcarbamate, CREB-binding protein, GLYCEROL, ... | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, Y, Zhang, C, Luo, G, Wu, X, Shen, H, Xu, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
7F7F
 
 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in yeast lipids with beryllium fluoride (resting state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2021-06-29 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7DRX
 
 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in 90PS with beryllium fluoride (E2P state) | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, Alkylphosphocholine resistance protein LEM3, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2020-12-30 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7DSH
 
 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in 90PS with AMPPCP (E1-ATP state) | | Descriptor: | Alkylphosphocholine resistance protein LEM3, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2020-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
7DSI
 
 | | Cryo-EM structure of Dnf1 from Saccharomyces cerevisiae in yeast lipids with AMPPCP ( resting state ) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkylphosphocholine resistance protein LEM3, ... | | Authors: | Xu, J, He, Y, Wu, X, Li, L. | | Deposit date: | 2020-12-31 | | Release date: | 2022-03-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Conformational changes of a phosphatidylcholine flippase in lipid membranes.
Cell Rep, 38, 2022
|
|
4LLW
 
 | | Crystal structure of Pertuzumab Clambda Fab with variable domain redesign (VRD2) at 1.95A | | Descriptor: | SULFATE ION, light chain Clambda, mutated Pertuzumab Fab heavy chain | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
