6PEW
 
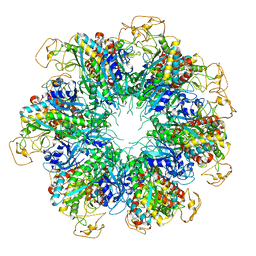 | |
6PEV
 
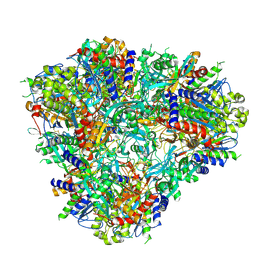 | |
3WX4
 
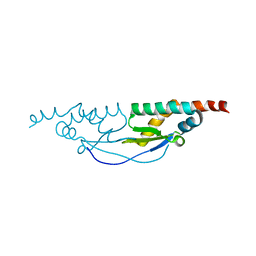 | |
6E10
 
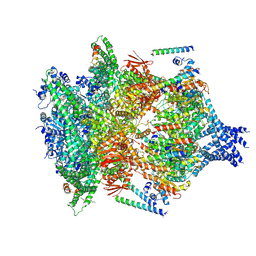 | | PTEX Core Complex in the Engaged (Extended) State | | Descriptor: | Endogenous cargo polypeptide, Exported protein 2, Heat shock protein 101, ... | | Authors: | Ho, C, Lai, M, Zhou, Z.H. | | Deposit date: | 2018-07-08 | | Release date: | 2018-08-22 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Malaria parasite translocon structure and mechanism of effector export.
Nature, 561, 2018
|
|
6E11
 
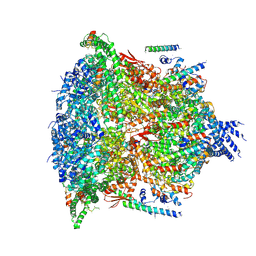 | | PTEX Core Complex in the Resetting (Compact) State | | Descriptor: | Endogenous cargo polypeptide, Exported protein 2, Heat shock protein 101, ... | | Authors: | Ho, C, Lai, M, Zhou, Z.H. | | Deposit date: | 2018-07-08 | | Release date: | 2018-08-22 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Malaria parasite translocon structure and mechanism of effector export.
Nature, 561, 2018
|
|
7D20
 
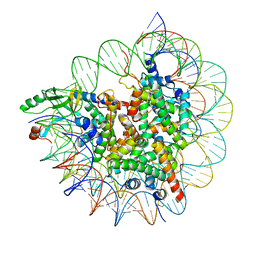 | | Cryo-EM structure of SET8-CENP-A-nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Ho, C.-H, Takizawa, Y, Kobayashi, W, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2020-09-15 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of nucleosomal histone H4 lysine 20 methylation by SET8 methyltransferase.
Life Sci Alliance, 4, 2021
|
|
7D1Z
 
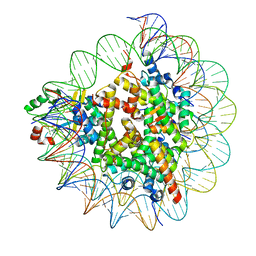 | | Cryo-EM structure of SET8-nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Ho, C.-H, Takizawa, Y, Kobayashi, W, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2020-09-15 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of nucleosomal histone H4 lysine 20 methylation by SET8 methyltransferase.
Life Sci Alliance, 4, 2021
|
|
1S68
 
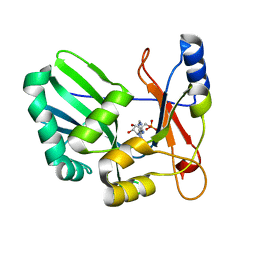 | | Structure and Mechanism of RNA Ligase | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA Ligase 2 | | Authors: | Ho, C.K, Wang, L.K, Lima, C.D, Shuman, S. | | Deposit date: | 2004-01-22 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of RNA ligase.
Structure, 12, 2004
|
|
4PDS
 
 | | Crystal structure of Rad53 kinase domain and SCD2 in complex with AMPPNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase RAD53 | | Authors: | Ho, C.S, Wybenga-Groot, L.E, Ceccarelli, D.F, Sicheri, F. | | Deposit date: | 2014-04-21 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of Rad53 kinase activation by dimerization and activation segment exchange.
Cell Signal., 26, 2014
|
|
7MRW
 
 | | Native RhopH complex of the malaria parasite Plasmodium falciparum | | Descriptor: | Cytoadherence linked asexual protein 3.1, High molecular weight rhoptry protein 2, High molecular weight rhoptry protein 3 | | Authors: | Ho, C.M, Jih, J, Lai, M, Li, X.R, Goldberg, D.E, Beck, J.R, Zhou, Z.H. | | Deposit date: | 2021-05-10 | | Release date: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Native structure of the RhopH complex, a key determinant of malaria parasite nutrient acquisition.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3KB2
 
 | | Crystal Structure of YorR protein in complex with phosphorylated GDP from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR256 | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE-5'-DIPHOSPHATE, MAGNESIUM ION, SPBc2 prophage-derived uncharacterized protein yorR | | Authors: | Forouhar, F, Friedman, D, Seetharaman, J, Janjua, J, Xiao, R, Cunningham, K, Ma, L, Ho, C, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-20 | | Release date: | 2009-10-27 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Northeast Structural Genomics Consortium Target SR256
To be Published
|
|
4LLD
 
 | | Structure of wild-type IgG1 antibody heavy chain constant domain 1 and light chain lambda constant domain (IgG1 CH1:Clambda) at 1.19A | | Descriptor: | Ig gamma-1 chain C region, Ig lambda-2 chain C region | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
4LLM
 
 | | Structure of redesigned IgG1 first constant and lambda domains (CH1:Clambda constant redesign 1, CRD1) at 1.75A | | Descriptor: | Ig gamma-1 chain C region, Ig lambda-2 chain C region | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
4LLY
 
 | | Crystal structure of Pertuzumab Clambda Fab with variable and constant domain redesigns (VRD2 and CRD2) at 1.6A | | Descriptor: | GLYCEROL, MAGNESIUM ION, light chain Clambda, ... | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
4LLU
 
 | | Structure of Pertuzumab Fab with light chain Clambda at 2.16A | | Descriptor: | ACETATE ION, Light chain CLAMBDA, PERTUZUMAB FAB Heavy chain, ... | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
4LLQ
 
 | | Structure of redesigned IgG1 first constant and lambda domains (CH1:Clambda constant redesign 2 beta, CRD2b) at 1.42A | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, mutated CH1, mutated light chain Clambda | | Authors: | Pustilnik, A, Lewis, S.M, Wu, X, Sereno, A, Huang, F, Guntas, G, Leaver-Fay, A, Smith, E.M, Ho, C, Hansen-Estruch, C, Chamberlain, A.K, Truhlar, S.M, Kuhlman, B, Demarest, S.J, Atwell, S. | | Deposit date: | 2013-07-09 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface.
Nat.Biotechnol., 32, 2014
|
|
2KEW
 
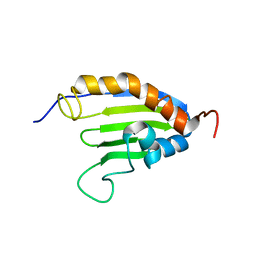 | | The solution structure of Bacillus subtilis SR211 START domain by NMR spectroscopy | | Descriptor: | Uncharacterized protein yndB | | Authors: | Mercier, K.A, Mueller, G.A, Powers, R, Acton, T.B, Ciano, M, Ho, C, Lui, J, Ma, L, Rost, B, Rossi, R, Xiao, R, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | (1)H, (13)C, and (15)N NMR assignments for the Bacillus subtilis yndB START domain.
Biomol.Nmr Assign., 3, 2009
|
|
1RVW
 
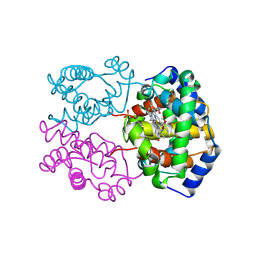 | | R STATE HUMAN HEMOGLOBIN [ALPHA V96W], CARBONMONOXY | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN, PHOSPHATE ION, ... | | Authors: | Puius, Y.A, Zou, M, Ho, N.T, Ho, C, Almo, S.C. | | Deposit date: | 1997-03-20 | | Release date: | 1998-03-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel water-mediated hydrogen bonds as the structural basis for the low oxygen affinity of the blood substitute candidate rHb(alpha 96Val-->Trp).
Biochemistry, 37, 1998
|
|
3HD6
 
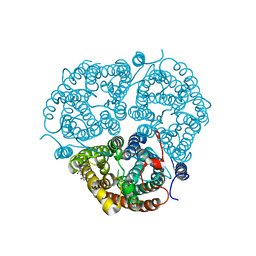 | | Crystal Structure of the Human Rhesus Glycoprotein RhCG | | Descriptor: | Ammonium transporter Rh type C, octyl beta-D-glucopyranoside | | Authors: | Gruswitz, F, Chaudhary, S, Ho, J.D, Pezeshki, B, Ho, C.-M, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2009-05-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Function of human Rh based on structure of RhCG at 2.1 A.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1VWT
 
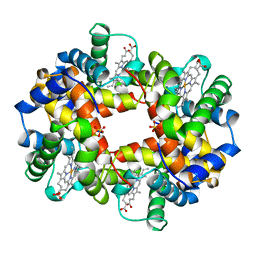 | | T STATE HUMAN HEMOGLOBIN [ALPHA V96W], ALPHA AQUOMET, BETA DEOXY | | Descriptor: | HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Puius, Y.A, Zou, M, Ho, N.T, Ho, C, Almo, S.C. | | Deposit date: | 1997-03-20 | | Release date: | 1998-03-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel water-mediated hydrogen bonds as the structural basis for the low oxygen affinity of the blood substitute candidate rHb(alpha 96Val-->Trp).
Biochemistry, 37, 1998
|
|
2KTE
 
 | | The solution structure of Bacillus subtilis, YndB, Northeast Structural Genomics Consoritum Target SR211 | | Descriptor: | Uncharacterized protein yndB | | Authors: | Mercier, K.A, Mueller, G.A, Powers, R, Acton, T.B, Ciano, M, Ho, C, Lui, J, Ma, L, Rost, B, Rossi, R, Montelione, G.T, Xiao, R, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and function of YndB, an AHSA1 protein from Bacillus subtilis.
Proteins, 78, 2010
|
|
8TPU
 
 | | Subtomogram averaged consensus structure of the malarial 80S ribosome in Plasmodium falciparum-infected human erythrocytes | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Anton, L, Cheng, W, Zhu, X, Ho, C.M. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Divergent translational landscape reflects adaptation to biased codon usage in malaria parasites
To Be Published
|
|
4CCL
 
 | | X-Ray structure of E. coli ycfD | | Descriptor: | 50S RIBOSOMAL PROTEIN L16 ARGININE HYDROXYLASE, MANGANESE (II) ION, SULFATE ION | | Authors: | McDonough, M.A, Ho, C.H, Kershaw, N.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-04-30 | | Last modified: | 2020-06-03 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
2FFG
 
 | | Novel x-ray structure of the YkuJ protein from Bacillus subtilis. Northeast Structural Genomics target SR360. | | Descriptor: | ykuJ | | Authors: | Kuzin, A.P, Abashidze, M, Forouhar, F, Vorobiev, S.M, Ho, C.K, Janjua, H, Cunningham, K, Conover, K, Ma, L.C, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-19 | | Release date: | 2005-12-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Novel x-ray structure of the YkuJ protein from Bacillus subtilis. Northeast Structural Genomics target SR360.
To be Published
|
|
2BDR
 
 | | Crystal Structure of the Putative Ureidoglycolate hydrolase PP4288 from Pseudomonas putida, Northeast Structural Genomics Target PpR49 | | Descriptor: | SODIUM ION, Ureidoglycolate hydrolase | | Authors: | Forouhar, F, Abashidze, M, Jayaraman, S, Ho, C.K, Conover, K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-20 | | Release date: | 2005-11-15 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Putative Ureidoglycolate hydrolase PP4288 from Pseudomonas putida, Northeast Structural Genomics Target PpR49
To be Published
|
|
