8KFY
 
 | | Gi bound CCR8 complex with nonpeptide agonist ZK 756326 | | Descriptor: | 2-[2-[4-[(3-phenoxyphenyl)methyl]piperazin-1-yl]ethoxy]ethanol, C-C chemokine receptor type 8,Fusion protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Jiang, S, Lin, X, Wu, L.J, Xu, F. | | Deposit date: | 2023-08-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Unveiling the structural mechanisms of nonpeptide ligand recognition and activation in human chemokine receptor CCR8.
Sci Adv, 10, 2024
|
|
7DRV
 
 | | Structural basis of SARS-CoV-2-closely-related bat coronavirus RaTG13 to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liu, K.F, Pan, X.Q, Li, L.J, Feng, Y, Meng, Y.M, Zhang, Y.F, Wu, L.L, Chen, Q, Zheng, A.Q, Song, C.L, Jia, Y.F, Niu, S, Qiao, C.P, Zhao, X, Ma, D.L, Ma, X.P, Tan, S.G, Qi, J.X, Gao, G.F, Wang, Q.H. | | Deposit date: | 2020-12-29 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Binding and molecular basis of the bat coronavirus RaTG13 virus to ACE2 in humans and other species.
Cell, 184, 2021
|
|
4GAW
 
 | | Crystal structure of active human granzyme H | | Descriptor: | CHLORIDE ION, Granzyme H, SULFATE ION | | Authors: | Wang, L, Li, Q, Wu, L, Zhang, K, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of SERPINB1 as a physiological inhibitor of human granzyme H
J.Immunol., 190, 2013
|
|
6AJH
 
 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with AU1235 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1-(2-adamantyl)-3-[2,3,4-tris(fluoranyl)phenyl]urea, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJI
 
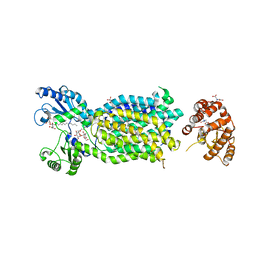 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with Rimonabant | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-N-(piperidin-1-yl)-1H-pyrazole-3-carboxamide, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJF
 
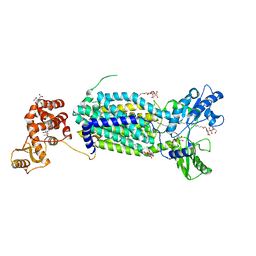 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Drug exporters of the RND superfamily-like protein,Endolysin, alpha-D-glucopyranosyl 6-O-dodecyl-alpha-D-glucopyranoside | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJG
 
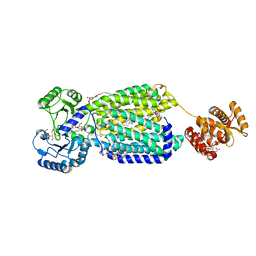 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with SQ109 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, DODECYL-BETA-D-MALTOSIDE, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
6AJJ
 
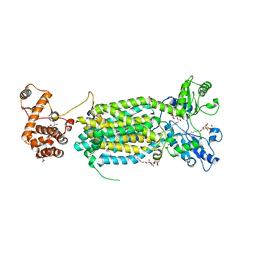 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with ICA38 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 4,6-difluoro-N-(spiro[5.5]undecan-3-yl)-1H-indole-2-carboxamide, Drug exporters of the RND superfamily-like protein,Endolysin, ... | | Authors: | Zhang, B, Li, J, Yang, X.L, Wu, L.J, Yang, H.T, Rao, Z.H. | | Deposit date: | 2018-08-27 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target.
Cell, 176, 2019
|
|
5ZOA
 
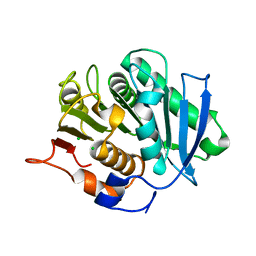 | |
7DH7
 
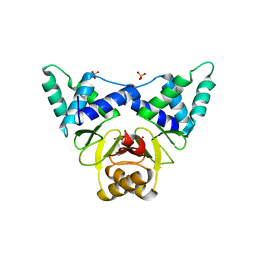 | | Crystal structure of apo XcZur | | Descriptor: | PHOSPHATE ION, Transcriptional regulator fur family, ZINC ION | | Authors: | Liu, F.M, Su, Z.H, Chen, P, Tian, X.L, Wu, L.J, Tang, D.J, Li, P.F, Deng, H.T, Tang, J.L, Ming, Z.H. | | Deposit date: | 2020-11-13 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for zinc-induced activation of a zinc uptake transcriptional regulator.
Nucleic Acids Res., 49, 2021
|
|
7CZE
 
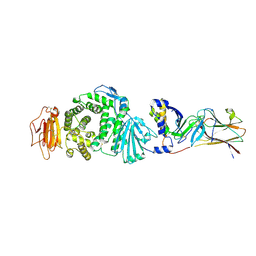 | | Crystal structure of Epstein-Barr virus (EBV) gHgL and in complex with the ligand binding domian (LBD) of EphA2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, Envelope glycoprotein L, ... | | Authors: | Su, C, Wu, L.L, Song, H, Chai, Y, Qi, J.X, Yan, J.H, Gao, G.F. | | Deposit date: | 2020-09-08 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis of EphA2 recognition by gHgL from gammaherpesviruses.
Nat Commun, 11, 2020
|
|
3O5X
 
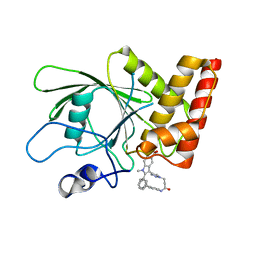 | | Crystal structure of the oncogenic tyrosine phosphatase SHP2 complexed with a salicylic acid-based small molecule inhibitor | | Descriptor: | 3-{1-[3-(biphenyl-4-ylamino)-3-oxopropyl]-1H-1,2,3-triazol-4-yl}-6-hydroxy-1-methyl-2-phenyl-1H-indole-5-carboxylic acid, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Zhang, Z.-Y, Zhang, X, He, Y, Liu, S, Yu, Z, Jiang, Z, Yang, Z, Dong, Y, Nabinger, S.C, Wu, L, Gunawan, A.M, Wang, L, Chan, R.J. | | Deposit date: | 2010-07-28 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Salicylic acid based small molecule inhibitor for the oncogenic Src homology-2 domain containing protein tyrosine phosphatase-2 (SHP2).
J.Med.Chem., 53, 2010
|
|
7DH8
 
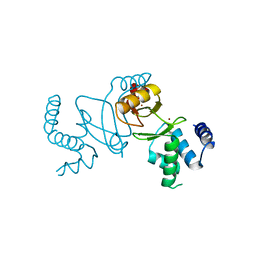 | | Crystal structure of holo XcZur | | Descriptor: | Transcriptional regulator fur family, ZINC ION | | Authors: | Liu, F.M, Su, Z.H, Chen, P, Tian, X.L, Wu, L.J, Tang, D.J, Li, P.F, Deng, H.T, Tang, J.L, Ming, Z.H. | | Deposit date: | 2020-11-13 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for zinc-induced activation of a zinc uptake transcriptional regulator.
Nucleic Acids Res., 49, 2021
|
|
6LCU
 
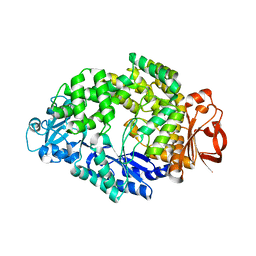 | |
4OHL
 
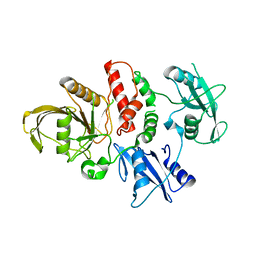 | | LEOPARD Syndrome-Associated SHP2/T468M mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Zhang, R.Y, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L, Liu, S, Zhang, Z.Y. | | Deposit date: | 2014-01-17 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of gain-of-function LEOPARD syndrome-associated SHP2 mutations.
Biochemistry, 53, 2014
|
|
4GA7
 
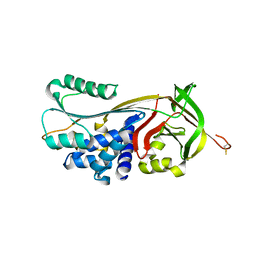 | | Crystal structure of human serpinB1 mutant | | Descriptor: | Leukocyte elastase inhibitor | | Authors: | Wang, L, Li, Q, Wu, L, Zhang, K, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of SERPINB1 as a physiological inhibitor of human granzyme H
J.Immunol., 190, 2013
|
|
4OHH
 
 | | LEOPARD Syndrome-Associated SHP2/Q506P mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Zhang, R.Y, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L, Liu, S, Zhang, Z.Y. | | Deposit date: | 2014-01-17 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of gain-of-function LEOPARD syndrome-associated SHP2 mutations.
Biochemistry, 53, 2014
|
|
1PA9
 
 | | Yersinia Protein-Tyrosine Phosphatase complexed with pNCS (Yop51,Pasteurella X,Ptpase,Yop51delta162) (Catalytic Domain, Residues 163-468) Mutant With Cys 235 Replaced By Arg (C235r) | | Descriptor: | N,4-DIHYDROXY-N-OXO-3-(SULFOOXY)BENZENAMINIUM, Protein-tyrosine phosphatase yopH | | Authors: | Sun, J.P, Wu, L, Fedorov, A.A, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2003-05-13 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Yersinia protein-tyrosine phosphatase YopH complexed
with a specific small molecule inhibitor
J.BIOL.CHEM., 278, 2003
|
|
7WD1
 
 | | Crystal structure of R14 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, R14, Spike protein S1, ... | | Authors: | Wang, Q.H, Gao, G.F, Qi, J.X, Su, C, Liu, H.H, Wu, L.L. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two pan-SARS-CoV-2 nanobodies and their multivalent derivatives effectively prevent Omicron infections in mice.
Cell Rep Med, 4, 2023
|
|
7WD2
 
 | | Crystal structure of S43 bound to SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Q.H, Gao, G.F, Qi, J.X, Su, C, Liu, H.H, Wu, L.L. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Two pan-SARS-CoV-2 nanobodies and their multivalent derivatives effectively prevent Omicron infections in mice.
Cell Rep Med, 4, 2023
|
|
5G0Q
 
 | | beta-glucuronidase with an activity-based probe (N-alkyl cyclophellitol aziridine) bound | | Descriptor: | (1R,2S,3R,4S,5S,6R)-7-(8-AZIDOOCTYL)-3,4,5-TRIHYDROXY-, BETA-GLUCURONIDASE, GLYCEROL | | Authors: | Jin, Y, Wu, L, Jiang, J.B, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2016-03-22 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
3TJU
 
 | | Crystal structure of human granzyme H with an inhibitor | | Descriptor: | Ac-PTSY-CMK inhibitor, Granzyme H, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
7XK2
 
 | | Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, DiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
3TK9
 
 | | Crystal structure of human granzyme H | | Descriptor: | Granzyme H, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
3TJV
 
 | | Crystal structure of human granzyme H with a peptidyl substrate | | Descriptor: | Granzyme H, PTSYAGDDSG, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
