6XYO
 
 | | Multiple system atrophy Type I alpha-synuclein filament | | Descriptor: | Alpha-synuclein | | Authors: | Schweighauser, M, Shi, Y, Tarutani, A, Kametani, F, Murzin, A.G, Ghetti, B, Matsubara, T, Tomita, T, Ando, T, Hasegawa, K, Murayama, S, Yoshida, M, Hasegawa, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of alpha-synuclein filaments from multiple system atrophy.
Nature, 585, 2020
|
|
6XYP
 
 | | Multiple system atrophy Type II-1 alpha-synuclein filament | | Descriptor: | Alpha-synuclein | | Authors: | Schweighauser, M, Shi, Y, Tarutani, A, Kametani, F, Murzin, A.G, Ghetti, B, Matsubara, T, Tomita, T, Ando, T, Hasegawa, K, Murayama, S, Yoshida, M, Hasegawa, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structures of alpha-synuclein filaments from multiple system atrophy.
Nature, 585, 2020
|
|
7QGV
 
 | | Solid-state NMR structure of Teixobactin-Lipid II. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, 3-methylbut-2-en-1-ol, Lipid II, ... | | Authors: | Weingarth, M.H, Shukla, R. | | Deposit date: | 2021-12-10 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-15 | | Method: | SOLID-STATE NMR | | Cite: | Teixobactin kills bacteria by a two-pronged attack on the cell envelope.
Nature, 608, 2022
|
|
6XYQ
 
 | | Multiple system atrophy Type II-2 alpha-synuclein filament | | Descriptor: | Alpha-synuclein | | Authors: | Schweighauser, M, Shi, Y, Tarutani, A, Kametani, F, Murzin, A.G, Ghetti, B, Matsubara, T, Tomita, T, Ando, T, Hasegawa, K, Murayama, S, Yoshida, M, Hasegawa, M, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2020-01-30 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structures of alpha-synuclein filaments from multiple system atrophy.
Nature, 585, 2020
|
|
6YFY
 
 | | Solid-state NMR structure of the D-Arg4,L10-teixobactin - Lipid II complex in lipid bilayers. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-N-acetyl-alpha-muramic acid, 3-methylbut-2-en-1-ol, D-Arg4,Leu10-Teixobactin, ... | | Authors: | Weingarth, M.H, Shukla, R. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-15 | | Method: | SOLID-STATE NMR | | Cite: | Mode of action of teixobactins in cellular membranes.
Nat Commun, 11, 2020
|
|
8V2F
 
 | | Crystal structure of IRAK4 kinase domain with compound 9 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 2024
|
|
8V1O
 
 | | Crystal structure of IRAK4 kinase domain with compound 4 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 2024
|
|
8V2L
 
 | | Crystal structure of IRAK4 kinase domain with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, Interleukin-1 receptor-associated kinase 4, N-{2-[4-(hydroxymethyl)phenyl]-6-(2-hydroxypropan-2-yl)-2H-indazol-5-yl}-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 2024
|
|
8R1A
 
 | | Model of the membrane-bound GBP1 oligomer | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, Guanylate binding protein 1, ... | | Authors: | Weismehl, M, Chu, X, Kutsch, M, Lauterjung, P, Herrmann, C, Kudryashev, M, Daumke, O. | | Deposit date: | 2023-11-01 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (26.799999 Å) | | Cite: | Structural insights into the activation mechanism of antimicrobial GBP1.
Embo J., 43, 2024
|
|
8P34
 
 | | Tau filaments extracted from human brain with the DeltaK281 mutation in MAPT | | Descriptor: | Microtubule-associated protein tau | | Authors: | Schweighauser, M, Garringer, H.J, Klingstedt, T, Masuda-Suzukake, M, Murrell, J.R, Vidal, R, Scheres, S.H.W, Goedert, M, Ghetti, B, Newell, K.L. | | Deposit date: | 2023-05-17 | | Release date: | 2023-07-05 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Mutation ∆K281 in MAPT causes Pick's disease.
Acta Neuropathol, 146, 2023
|
|
8Q92
 
 | | P301S Tau Filaments from the Brains of PS19 Transgenic Mouse Line | | Descriptor: | Microtubule-associated protein tau | | Authors: | Schweighauser, M, Murzin, A.G, Macdonald, J, Lavenir, I, Crowther, R.A, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-19 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Cryo-EM structures of tau filaments from the brains of mice transgenic for human mutant P301S Tau.
Acta Neuropathol Commun, 11, 2023
|
|
8Q96
 
 | | P301S Tau Filaments from the Brains of Tg2541 Transgenic Mouse Line | | Descriptor: | Isoform Tau-E of Microtubule-associated protein tau, Unknown protein | | Authors: | Schweighauser, M, Murzin, A.G, Macdonald, J, Lavenir, I, Crowther, R.A, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2023-08-19 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Cryo-EM structures of tau filaments from the brains of mice transgenic for human mutant P301S Tau.
Acta Neuropathol Commun, 11, 2023
|
|
6X4X
 
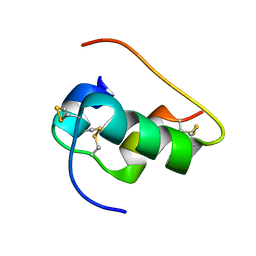 | | B24Y DKP insulin | | Descriptor: | Insulin, Insulin chain A | | Authors: | Weiss, M.A, Yang, Y. | | Deposit date: | 2020-05-24 | | Release date: | 2020-08-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Evolution of insulin at the edge of foldability and its medical implications.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4BTS
 
 | | THE CRYSTAL STRUCTURE OF THE EUKARYOTIC 40S RIBOSOMAL SUBUNIT IN COMPLEX WITH EIF1 AND EIF1A | | Descriptor: | 18S ribosomal RNA, 40S RIBOSOMAL PROTEIN RACK1, 40S RIBOSOMAL PROTEIN RPS10E, ... | | Authors: | Weisser, M, Voigts-Hoffmann, F, Rabl, J, Leibundgut, M, Ban, N. | | Deposit date: | 2013-06-19 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.703 Å) | | Cite: | The crystal structure of the eukaryotic 40S ribosomal subunit in complex with eIF1 and eIF1A.
Nat. Struct. Mol. Biol., 20, 2013
|
|
1OT8
 
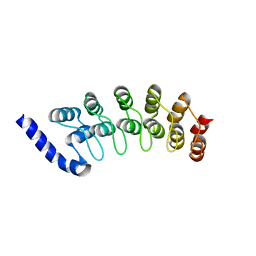 | | Structure of the Ankyrin Domain of the Drosophila Notch Receptor | | Descriptor: | MAGNESIUM ION, Neurogenic locus Notch protein | | Authors: | Zweifel, M.E, Leahy, D.J, Hughson, F.M, Barrick, D. | | Deposit date: | 2003-03-21 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and stability of the ankyrin domain of the Drosophila Notch receptor
Protein Sci., 12, 2003
|
|
1JCA
 
 | | Non-standard Design of Unstable Insulin Analogues with Enhanced Activity | | Descriptor: | ZINC ION, insulin a, insulin b | | Authors: | Weiss, M.A, Wan, Z, Zhao, M, Chu, Y.-C, Nakagawa, S.H, Burke, G.T, Jia, W, Hellmich, R, Katsoyannis, P.G. | | Deposit date: | 2001-06-08 | | Release date: | 2001-06-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-standard insulin design: structure-activity relationships at the periphery of the insulin receptor.
J.Mol.Biol., 315, 2002
|
|
1DTP
 
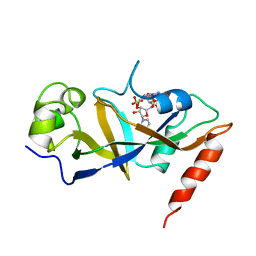 | |
1F13
 
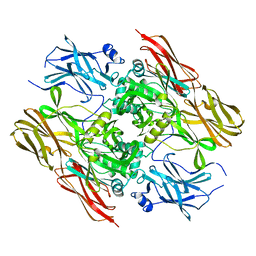 | |
3JSD
 
 | | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Weiss, M.A, Wan, Z.L, Dodson, E.J, Liu, M, Xu, B, Hua, Q.X, Turkenburg, M, Whittingham, J, Nakagawa, S.H, Huang, K, Hu, S.Q, Jia, W.H, Wang, S.H, Brange, J, Whittaker, J, Arvan, P, Katsoyannis, P.G, Dodson, G.G. | | Deposit date: | 2009-09-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insulin's biosynthesis and activity have opposing structural requirements: a new factor in neonatal diabetes mellitus
To be Published
|
|
1ZV1
 
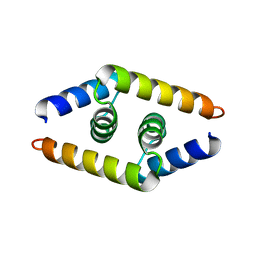 | | Crystal structure of the dimerization domain of doublesex protein from D. melanogaster | | Descriptor: | Doublesex protein | | Authors: | Weiss, M.A, Bayrer, J.R, Wan, Z, Li, B, Phillips, N.B. | | Deposit date: | 2005-06-01 | | Release date: | 2005-08-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dimerization of doublesex is mediated by a cryptic ubiquitin-associated domain fold: implications for sex-specific gene regulation
J.Biol.Chem., 280, 2005
|
|
1P23
 
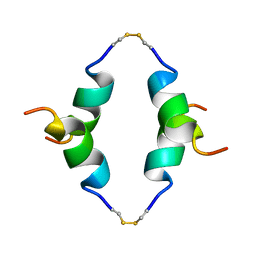 | | STRUCTURE OF THE DIMERIZED CYTOPLASMIC DOMAIN OF P23 IN SOLUTION, NMR, 10 STRUCTURES | | Descriptor: | TRANSMEMBRANE PROTEIN TMP21 PRECURSOR | | Authors: | Weidler, M, Reinhard, C, Wieland, F.T, Roesch, P. | | Deposit date: | 1998-11-17 | | Release date: | 2000-06-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of the cytoplasmic domain of p23 in solution: implications for the formation of COPI vesicles.
Biochem.Biophys.Res.Commun., 271, 2000
|
|
5RC0
 
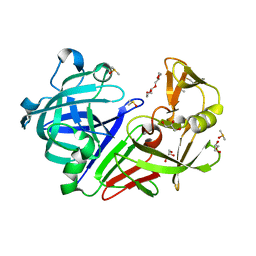 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library E06a | | Descriptor: | (3-endo)-8-benzyl-8-azabicyclo[3.2.1]octan-3-ol, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RCD
 
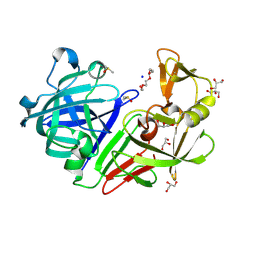 | | PanDDA analysis group deposition -- Endothiapepsin changed state model for fragment F2X-Entry Library H03a | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RCT
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 15 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5RD9
 
 | | PanDDA analysis group deposition -- Endothiapepsin ground state model 31 | | Descriptor: | Endothiapepsin | | Authors: | Weiss, M.S, Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G. | | Deposit date: | 2020-03-24 | | Release date: | 2020-06-03 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
