2OO1
 
 | | Crystal structure of the Bromo domain 2 of human Bromodomain containing protein 3 (BRD3) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Bromodomain-containing protein 3, ... | | Authors: | Filippakopoulos, P, Bullock, A, Papagrigoriou, E, Keates, T, Cooper, C, Smee, C, Ugochukwu, E, Debreczeni, J, von Delft, F, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
2OOQ
 
 | | Crystal Structure of the Human Receptor Phosphatase PTPRT | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Receptor-type tyrosine-protein phosphatase T, ... | | Authors: | Ugochukwu, E, Alfano, I, Barr, A, Keates, T, Eswaran, J, Salah, E, Savitsky, P, Bunkoczi, G, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-26 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
6FCX
 
 | | Structure of human 5,10-methylenetetrahydrofolate reductase (MTHFR) | | Descriptor: | CITRIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Methylenetetrahydrofolate reductase, ... | | Authors: | Kopec, J, Bezerra, G.A, Oberholzer, A.E, Rembeza, E, Sorrell, F.J, Chalk, R, Borkowska, O, Ellis, K, Kupinska, K, Krojer, T, Burgess-Brown, N, Von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Froese, D.S, Baumgartner, M, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-21 | | Release date: | 2018-05-16 | | Last modified: | 2018-07-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the regulation of human 5,10-methylenetetrahydrofolate reductase by phosphorylation and S-adenosylmethionine inhibition.
Nat Commun, 9, 2018
|
|
6FAE
 
 | | The Sec7 domain of IQSEC2 (Brag1) in complex with the small GTPase Arf1 | | Descriptor: | 1,2-ETHANEDIOL, ADP-ribosylation factor 1, IQ motif and SEC7 domain-containing protein 2 | | Authors: | Gray, J, Krojer, T, Fairhead, M, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P, von Delft, F. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Targeting the Small GTPase Superfamily through their Regulatory Proteins.
Angew.Chem.Int.Ed.Engl., 2019
|
|
6GPJ
 
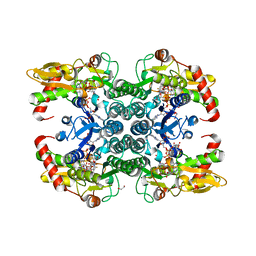 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase in complex with GDP-4F-Man | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, GDP-mannose 4,6 dehydratase, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
6H3A
 
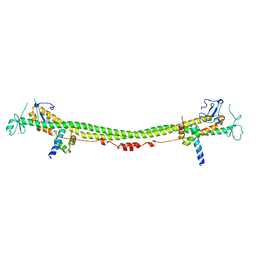 | | Crystal structure of the KAP1 RBCC domain in complex with the SMARCAD1 CUE1 domain. | | Descriptor: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1, Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Newman, J.A, Aitkenhead, H, Lim, M, Williams, H.L, Svejstrup, J.Q, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-07-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (5.505 Å) | | Cite: | A Ubiquitin-Binding Domain that Binds a Structural Fold Distinct from that of Ubiquitin.
Structure, 27, 2019
|
|
6GPK
 
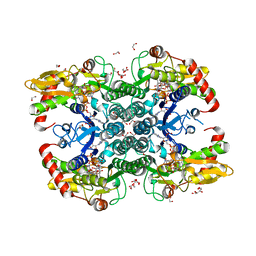 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase (E157Q) in complex with GDP-Man | | Descriptor: | 1,2-ETHANEDIOL, GDP-mannose 4,6 dehydratase, GLYCEROL, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
6GPL
 
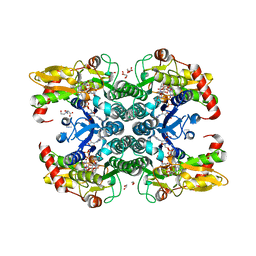 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase in complex with GDP-4k6d-Man | | Descriptor: | 1,2-ETHANEDIOL, BICINE, GDP-mannose 4,6 dehydratase, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
2X47
 
 | | Crystal structure of human MACROD1 | | Descriptor: | MACRO DOMAIN-CONTAINING PROTEIN 1, SULFATE ION | | Authors: | Vollmar, M, Phillips, C, Mehrotra, P.V, Ahel, I, Krojer, T, Yue, W, Ugochukwu, E, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Gileadi, O. | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of Macro Domain Proteins as Novel O-Acetyl-Adp-Ribose Deacetylases.
J.Biol.Chem., 286, 2011
|
|
2WO6
 
 | | Human Dual-Specificity Tyrosine-Phosphorylation-Regulated Kinase 1A in complex with a consensus substrate peptide | | Descriptor: | ARTIFICIAL CONSENSUS SEQUENCE, CHLORIDE ION, DUAL SPECIFICITY TYROSINE-PHOSPHORYLATION- REGULATED KINASE 1A, ... | | Authors: | Roos, A.K, Soundararajan, M, Elkins, J.M, Fedorov, O, Eswaran, J, Phillips, C, Pike, A.C.W, Ugochukwu, E, Muniz, J.R.C, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Wikstrom, M, Edwards, A, Bountra, C, Knapp, S. | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Down Syndrome Kinases, Dyrks, Reveal Mechanisms of Kinase Activation and Substrate Recognition.
Structure, 21, 2013
|
|
2WZ1
 
 | | STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN SOLUBLE GUANYLATE CYCLASE 1 BETA 3. | | Descriptor: | 1,2-ETHANEDIOL, GUANYLATE CYCLASE SOLUBLE SUBUNIT BETA-1 | | Authors: | Allerston, C.K, Cooper, C.D.O, Muniz, J, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2009-11-23 | | Release date: | 2009-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structures of the Catalytic Domain of Human Soluble Guanylate Cyclase.
Plos One, 8, 2013
|
|
2XML
 
 | | Crystal structure of human JMJD2C catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, LYSINE-SPECIFIC DEMETHYLASE 4C, ... | | Authors: | Yue, W.W, Gileadi, C, Krojer, T, Pike, A.C.W, von Delft, F, Ng, S, Carpenter, L, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2010-07-28 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Evolutionary Basis for the Dual Substrate Selectivity of Human Kdm4 Histone Demethylase Family.
J.Biol.Chem., 286, 2011
|
|
1T2A
 
 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase | | Descriptor: | GDP-mannose 4,6 dehydratase, GUANOSINE-5'-DIPHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Walker, J.R, Vedadi, M, Sharma, S, Houston, S, Wasney, G, Loppnau, P, Sundstrom, M, Arrowsmith, C, Edwards, A, Oppermann, U. | | Deposit date: | 2004-04-20 | | Release date: | 2004-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure and Biophysical Characterization of Human GDP-D-mannose 4,6-dehydratase
To be Published
|
|
2YPR
 
 | | Crystal structure of the DNA binding ETS domain of human protein FEV | | Descriptor: | GLYCEROL, PROTEIN FEV | | Authors: | Allerston, C.K, Cooper, C, Vollmar, M, Krojer, T, von Delft, F, Weigelt, J, Arrowsmith, C.H, Bountra, C, Edwards, A, Gileadi, O. | | Deposit date: | 2012-10-31 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structures of the Ets Domains of Transcription Factors Etv1, Etv4, Etv5 and Fev: Determinants of DNA Binding and Redox Regulation by Disulfide Bond Formation.
J.Biol.Chem., 290, 2015
|
|
4FLP
 
 | | Crystal Structure of the first bromodomain of human BRDT in complex with the inhibitor JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, Bromodomain testis-specific protein, POTASSIUM ION | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Canning, P, Muniz, J, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Bradner, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-15 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Small-Molecule Inhibition of BRDT for Male Contraception.
Cell(Cambridge,Mass.), 150, 2012
|
|
4HBV
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolin ligand | | Descriptor: | 1,2-ETHANEDIOL, 6-bromo-3-methyl-3,4-dihydroquinazolin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
4HBX
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolin ligand | | Descriptor: | 3-methyl-6-(pyrrolidin-1-ylsulfonyl)-3,4-dihydroquinazolin-2(1H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
4GPJ
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a isoxazolylbenzimidazole ligand | | Descriptor: | (1R)-6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-phenyl-2,3-dihydro-1H-inden-1-ol, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Heightman, T.D, Brennan, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The design and synthesis of 5- and 6-isoxazolylbenzimidazoles as selective inhibitors of the BET bromodomains.
Medchemcomm, 4, 2013
|
|
4HBW
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazoline ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-ethyl-3-methyl-2-oxo-1,2,3,4-tetrahydroquinazoline-6-sulfonamide, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
4HBY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolin ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-2-oxo-N-phenyl-1,2,3,4-tetrahydroquinazoline-6-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
5O1O
 
 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain with proline bound. | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, mitochondrial, ... | | Authors: | Kopec, J, Rembeza, E, Pena, I.A, Mathea, S, Velupillai, S, Strain-Damerell, C, Goubin, S, Kupinska, K, Talon, R, Collins, P, Krojer, T, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, von Delft, F, Arruda, P, Yue, W.W. | | Deposit date: | 2017-05-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain with proline bound.
To Be Published
|
|
5O1P
 
 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase. | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, mitochondrial, ... | | Authors: | Kopec, J, Rembeza, E, Pena, I.A, Williams, E, Velupillai, S, Kupinska, K, Strain-Damerell, C, Goubin, S, Talon, R, Collins, P, Krojer, T, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, von Delft, F, Arruda, P, Yue, W.W. | | Deposit date: | 2017-05-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase.
To Be Published
|
|
2JAV
 
 | | Human Kinase with pyrrole-indolinone ligand | | Descriptor: | 5-[(Z)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Pike, A.C.W, Rellos, P, Das, S, Fedorov, O, Papagrigoriou, E, Debreczeni, J.E, Turnbull, A.P, Gorrec, F, Bray, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A, Weigelt, J, von Delft, F, Knapp, S. | | Deposit date: | 2006-11-30 | | Release date: | 2006-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Regulation of the Human Nek2 Centrosomal Kinase
J.Biol.Chem., 282, 2007
|
|
5O1N
 
 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain with N-[(2S)-2-Pyrrolidinylmethyl]-trifluoromethanesulfonamide bound | | Descriptor: | 1,1,1-tris(fluoranyl)-~{N}-[[(2~{S})-pyrrolidin-2-yl]methyl]methanesulfonamide, 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, ... | | Authors: | Kopec, J, Rembeza, E, Pena, I.A, Strain-Damerell, C, Goubin, S, Sethi, R, Velupillai, S, Talon, R, Collins, P, Krojer, T, McLaughlin, M, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, von Delft, F, Arruda, P, Yue, W.W. | | Deposit date: | 2017-05-18 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain with N-[(2S)-2-Pyrrolidinylmethyl]-trifluoromethanesulfonamide bound
To Be Published
|
|
2JII
 
 | | Structure of vaccinia related kinase 3 | | Descriptor: | 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE VRK3 MOLECULE: VACCINIA RELATED KINASE 3 | | Authors: | Bunkoczi, G, Eswaran, J, Pike, A.C.W, Uppenberg, J, Ugochukwu, E, von Delft, F, Cooper, C, Salah, E, Savitsky, P, Burgess-Brown, N, Keates, T, Fedorov, O, Sobott, F, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Knapp, S. | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site.
Structure, 17, 2009
|
|
