7U4L
 
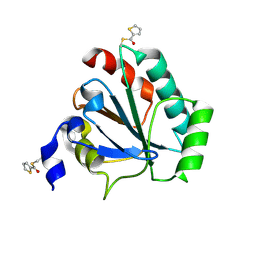 | | Crystal structure of human GPX4-U46C in complex with MAC-5576 | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase, thiophene-2-carbaldehyde | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4K
 
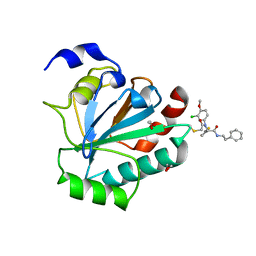 | | Crystal structure of human GPX4-U46C-R152H in complex with ML162 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-(3-chloro-4-methoxyphenyl)-N-[(1R)-2-oxo-2-[(2-phenylethyl)amino]-1-(thiophen-2-yl)ethyl]acetamide, Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Xia, X, Soni, R.K, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4N
 
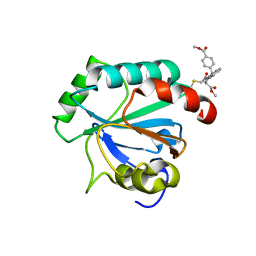 | | Crystal structure of human GPX4-U46C in complex with RSL3 | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase, methyl (1S,3R)-2-(chloroacetyl)-1-[4-(methoxycarbonyl)phenyl]-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole-3-carboxylate | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Xia, X, Soni, R.K, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4M
 
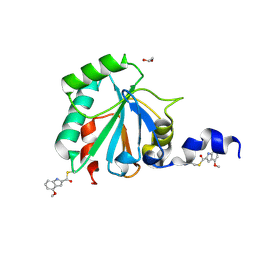 | | Crystal structure of human GPX4-U46C in complex with LOC1886 | | Descriptor: | 1,2-ETHANEDIOL, 4-methoxy-1H-indole-2-carbaldehyde, Phospholipid hydroperoxide glutathione peroxidase | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4J
 
 | | Crystal structure of human GPX4-U46C-R152H in complex with TMT10 | | Descriptor: | Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION, ~{N}-(3-chloranyl-4-methoxy-phenyl)ethanamide | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7U4I
 
 | | Crystal structure of human GPX4-U46C-R152H in complex with CDS9 | | Descriptor: | 2-bromo-N-[(thiophen-2-yl)methyl]acetamide, Phospholipid hydroperoxide glutathione peroxidase, THIOCYANATE ION | | Authors: | Forouhar, F, Liu, H, Lin, A.J, Wang, Q, Polychronidou, V, Soni, R.K, Xia, X, Stockwell, B.R. | | Deposit date: | 2022-02-28 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Small-molecule allosteric inhibitors of GPX4.
Cell Chem Biol, 29, 2022
|
|
7EN0
 
 | | Structure and Activity of SLAC1 Channels for Stomatal Signaling in Leaves | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, SLow Anion Channel 1, SPHINGOSINE | | Authors: | Deng, Y, Kashtoh, H, Wang, Q, Zhen, G, Li, Q, Tang, L, Gao, H, Zhang, C, Qin, L, Su, M, Li, F, Huang, X, Wang, Y, Xie, Q, Clarke, O.B, Hendrickson, W.A, Chen, Y. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and activity of SLAC1 channels for stomatal signaling in leaves.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BTF
 
 | | SARS-CoV-2 RNA-dependent RNA polymerase in complex with cofactors in reduced condition | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
2KDF
 
 | | NMR structure of minor S5a (196-306):K48 linked diubiquitin species | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4, Ubiquitin | | Authors: | Zhang, N, Wang, Q, Ehlinger, A, Randles, L, Lary, J.W, Kang, Y, Haririnia, A, Cole, J.L, Fushman, D, Walters, K.J. | | Deposit date: | 2009-01-06 | | Release date: | 2009-09-01 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13.
Mol.Cell, 35, 2009
|
|
2KDE
 
 | | NMR structure of major S5a (196-306):K48 linked diubiquitin species | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4, Ubiquitin | | Authors: | Zhang, N, Wang, Q, Ehlinger, A, Randles, L, Lary, J.W, Kang, Y, Haririnia, A, Cole, J.L, Fushman, D, Walters, K.J. | | Deposit date: | 2009-01-06 | | Release date: | 2009-09-01 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13.
Mol.Cell, 35, 2009
|
|
7BVE
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbC2-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, Ethambutol, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-01 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVC
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVG
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with di-arabinose. | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
5VMG
 
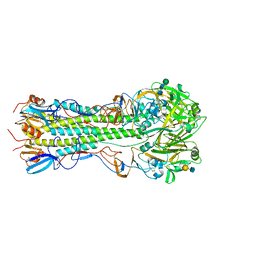 | | Influenza hemagglutinin H1 mutant DH1E in complex with 6'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Ni, F, Kondrashkina, E, Wang, Q. | | Deposit date: | 2017-04-27 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Determinant of receptor-preference switch in influenza hemagglutinin.
Virology, 513, 2017
|
|
5X58
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5B
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 2 | | Descriptor: | Spike glycoprotein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X59
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, three-fold symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5C
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 1 | | Descriptor: | S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5VMC
 
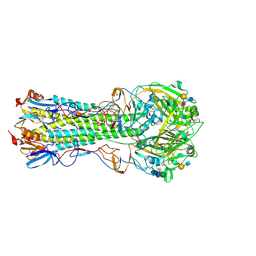 | | Influenza hemagglutinin H1 mutant DH1 in complex with 6'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Ni, F, Kondrashkina, E, Wang, Q. | | Deposit date: | 2017-04-27 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Determinant of receptor-preference switch in influenza hemagglutinin.
Virology, 513, 2017
|
|
5X5F
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 2 | | Descriptor: | S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2017-05-24 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
6LNI
 
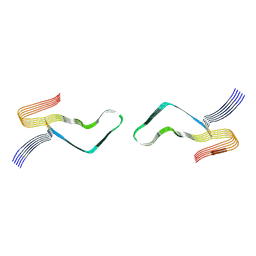 | | Cryo-EM structure of amyloid fibril formed by full-length human prion protein | | Descriptor: | Major prion protein | | Authors: | Wang, L.Q, Zhao, K, Yuan, H.Y, Wang, Q, Guan, Z.Y, Tao, J, Li, X.N, Hao, M.M, Chen, J, Zhang, D.L, Zhu, H.L, Yin, P, Liu, C, Liang, Y. | | Deposit date: | 2019-12-30 | | Release date: | 2020-06-10 | | Last modified: | 2020-06-24 | | Method: | ELECTRON MICROSCOPY (2.702 Å) | | Cite: | Cryo-EM structure of an amyloid fibril formed by full-length human prion protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6LUM
 
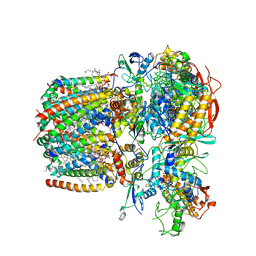 | | Structure of Mycobacterium smegmatis succinate dehydrogenase 2 | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ... | | Authors: | Gao, Y, Gong, H, Zhou, X, Xiao, Y, Wang, W, Ji, W, Wang, Q, Rao, Z. | | Deposit date: | 2020-01-29 | | Release date: | 2020-05-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of trimeric Mycobacterium smegmatis succinate dehydrogenase with a membrane-anchor SdhF.
Nat Commun, 11, 2020
|
|
7BVH
 
 | | Crystal structure of arabinosyltransferase EmbC2-AcpM2 complex from Mycobacterium smegmatis complexed with di-arabinose | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, Meromycolate extension acyl carrier protein, ... | | Authors: | Zhao, Y, Zhang, L, Wu, L.J, Wang, Q, Li, J, Besra, G.S, Rao, Z.H. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
5VMF
 
 | | Influenza hemagglutinin H1 mutant DH1D in complex with 6'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Ni, F, Kondrashkina, E, Wang, Q. | | Deposit date: | 2017-04-27 | | Release date: | 2017-11-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Determinant of receptor-preference switch in influenza hemagglutinin.
Virology, 513, 2017
|
|
7BVF
 
 | | Cryo-EM structure of Mycobacterium tuberculosis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, CARDIOLIPIN, Ethambutol, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
