2JLZ
 
 | | Dengue virus 4 NS3 helicase in complex with ssRNA and ADP | | Descriptor: | 5'-R(*AP*GP*AP*CP*UP*AP*AP*CP*AP*AP*CP*U)-3', ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
2JLY
 
 | | Dengue virus 4 NS3 helicase in complex with ssRNA and ADP-Phosphate | | Descriptor: | 5'-R(*AP*GP*AP*CP*UP*AP*AP*CP*AP*AP*CP*U)-3', ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
2JLX
 
 | | Dengue virus 4 NS3 helicase in complex with ssRNA and ADP-Vanadate | | Descriptor: | 5'-R(*AP*GP*AP*CP*UP*AP*AP*CP*AP*AP*CP*U)-3', ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
2JLU
 
 | | Dengue virus 4 NS3 helicase in complex with ssRNA | | Descriptor: | 5'-R(*AP*GP*AP*CP*UP*AP*AP*CP*AP*AP*CP*U)-3', GLYCEROL, SERINE PROTEASE SUBUNIT NS3 | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
3FMW
 
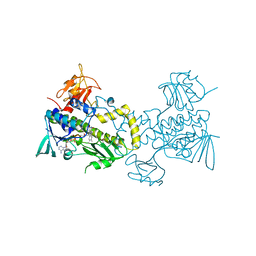 | | The crystal structure of MtmOIV, a Baeyer-Villiger monooxygenase from the mithramycin biosynthetic pathway in Streptomyces argillaceus. | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Oxygenase | | Authors: | Noinaj, N, Beam, M.P, Wang, C, Rohr, J. | | Deposit date: | 2008-12-22 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystal structure of Baeyer-Villiger monooxygenase MtmOIV, the key enzyme of the mithramycin biosynthetic pathway .
Biochemistry, 48, 2009
|
|
7XYR
 
 | |
8Q5Z
 
 | |
7VG8
 
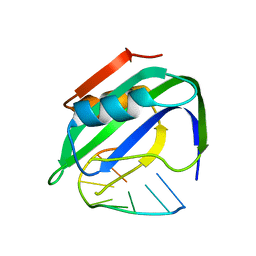 | | TYLCV Rep-DNA | | Descriptor: | DNA (5'-((DT)P*(DA)P*(DA)P*(DT)P*(DA)P*(DT)P*(DT)P*(DA)*(DC)P*)-3'), MANGANESE (II) ION, Replication-associated protein | | Authors: | Zhang, S.Y, Wang, C.N. | | Deposit date: | 2021-09-14 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | structure of TYLCV Rep complexed with ssDNA
To Be Published
|
|
2JQB
 
 | | Solution structure of a novel D-amiNo acid containing conopeptide, conomarphin at pH 5 | | Descriptor: | Conomarphin | | Authors: | Huang, F, Du, W, Han, Y, Wang, C, Chi, C. | | Deposit date: | 2007-05-31 | | Release date: | 2008-06-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel D-amiNo acid containing conopeptide, conomarphin at pH 5
To be Published
|
|
7EVJ
 
 | | Crystal structure of CBP bromodomain liganded with 9c | | Descriptor: | 3-acetyl-1-((3-(1-cyclopropyl-1H-pyrazol-4-yl)-2-fluoro-5-(hydroxymethyl)phenyl)carbamoyl)indolizin-7-yl dimethylcarbamate, CREB-binding protein, GLYCEROL, ... | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, Y, Zhang, C, Luo, G, Wu, X, Shen, H, Xu, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
7XIJ
 
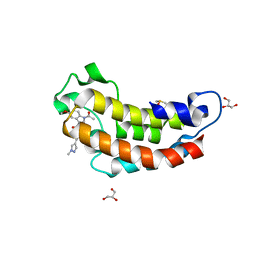 | | Crystal structure of CBP bromodomain liganded with Y08175 | | Descriptor: | 3-[(1-ethanoyl-5-methoxy-indol-3-yl)carbonylamino]-4-fluoranyl-5-(1-methylpyrazol-4-yl)benzoic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, Y, Zhang, C, Luo, G, Wu, X, Shen, H, Xu, Y. | | Deposit date: | 2022-04-13 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of CBP bromodomain liganded with Y08175
To Be Published
|
|
7XNE
 
 | | Crystal structure of CBP bromodomain liganded with Y08284 | | Descriptor: | CREB-binding protein, GLYCEROL, N-[3-(1-cyclopropylpyrazol-4-yl)-2-fluoranyl-5-[(1S)-1-oxidanylethyl]phenyl]-3-ethanoyl-7-methoxy-indolizine-1-carboxamide | | Authors: | Xiang, Q, Wang, C, Wu, T, Zhang, C, Hu, Q, Luo, G, Hu, J, Zhuang, X, Zou, L, Shen, H, Wu, X, Zhang, Y, Kong, X, Xu, Y. | | Deposit date: | 2022-04-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 1-(Indolizin-3-yl)ethan-1-ones as CBP Bromodomain Inhibitors for the Treatment of Prostate Cancer.
J.Med.Chem., 65, 2022
|
|
6N9O
 
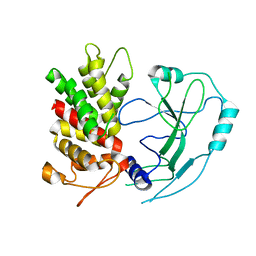 | | Crystal structure of human GSDMD | | Descriptor: | Gasdermin-D | | Authors: | Liu, Z, Wang, C, Yang, J, Xiao, T.S. | | Deposit date: | 2018-12-03 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structures of the Full-Length Murine and Human Gasdermin D Reveal Mechanisms of Autoinhibition, Lipid Binding, and Oligomerization.
Immunity, 51, 2019
|
|
4APP
 
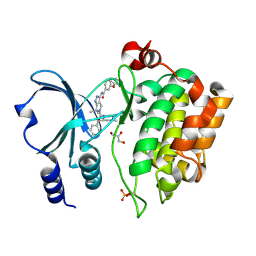 | | Crystal Structure of the Human p21-Activated Kinase 4 in Complex with (S)-N-(5-(3-benzyl-1-methylpiperazine-4-carbonyl)-6,6-dimethyl-1,4,5, 6-tetrahydropyrrolo(3,4-c)pyrazol-3-yl)-3-phenoxybenzamide | | Descriptor: | GLYCEROL, N-[6,6-dimethyl-5-[(2S)-4-methyl-2-(phenylmethyl)piperazin-1-yl]carbonyl-2,4-dihydropyrrolo[3,4-c]pyrazol-3-yl]-3-phenoxy-benzamide, SERINE/THREONINE-PROTEIN KINASE PAK 4 | | Authors: | Knighton, D.D, Deng, Y.L, Wang, C, Guo, C, McAlpine, I, Zhang, J, Kephart, S, Johnson, M.C, Li, H, Bouzida, D, Yang, A, Dong, L, Marakovits, J, Tikhe, J, Richardson, P, Guo, L.C, Kania, R, Edwards, M.P, Kraynov, E, Christensen, J, Piraino, J, Lee, J, Dagostino, E, Del-Carmen, C, Smeal, T, Murray, B.W. | | Deposit date: | 2012-04-04 | | Release date: | 2012-06-06 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Pyrroloaminopyrazoles as Novel Pak Inhibitors.
J.Med.Chem., 55, 2012
|
|
2MT8
 
 | |
2MY5
 
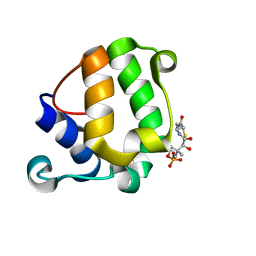 | | Solution Structure of KstB-PCP in kosinostatin biosynthesis | | Descriptor: | N~3~-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-(2-sulfanylethyl)-beta-alaninamide, Peptidyl carrier protein | | Authors: | Zhao, B, Lan, W, Wang, C, Tang, G, Cao, C. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | 1H and 15N Assigned Chemical Shifts for KstB-PCP
To be Published
|
|
2MY6
 
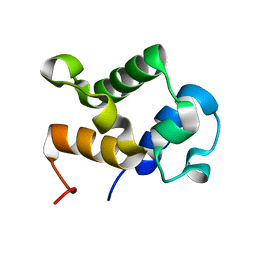 | |
6KZJ
 
 | | Crystal structure of Ankyrin B/NdeL1 complex | | Descriptor: | Ankyrin-2, Nuclear distribution protein nudE-like 1 | | Authors: | Ye, J, Li, J, Ye, F, Zhang, M, Zhang, Y, Wang, C. | | Deposit date: | 2019-09-24 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mechanistic insights into the interactions of dynein regulator Ndel1 with neuronal ankyrins and implications in polarity maintenance.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5Z6R
 
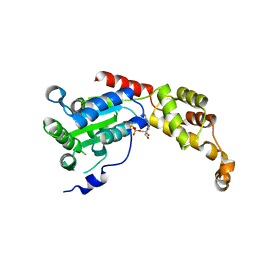 | | SPASTIN AAA WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Spastin | | Authors: | Lin, Z, Wang, C, Shen, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Aaa Family Protein Spastin Severs A Microtubule
To Be Published
|
|
5Z6Q
 
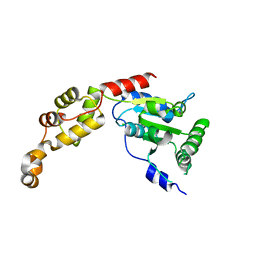 | | Crystal structure of AAA of Spastin | | Descriptor: | CHLORIDE ION, Spastin | | Authors: | Lin, Z, Wang, C, Shen, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2018-12-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The AAA protein spastin possesses two levels of basal ATPase activity
FEBS Lett., 592, 2018
|
|
2JLQ
 
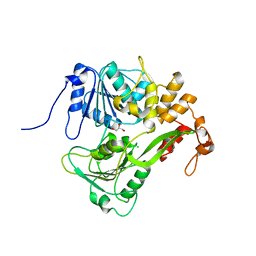 | | Dengue virus 4 NS3 helicase structure, apo enzyme. | | Descriptor: | CHLORIDE ION, GLYCEROL, SERINE PROTEASE SUBUNIT NS3 | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
2JLR
 
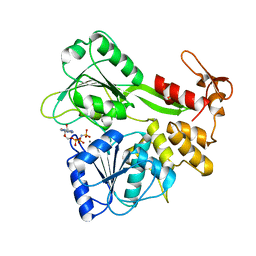 | | Dengue virus 4 NS3 helicase in complex with AMPPNP | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SERINE PROTEASE SUBUNIT NS3 | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
2JLS
 
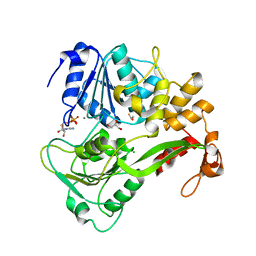 | | Dengue virus 4 NS3 helicase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
6NIV
 
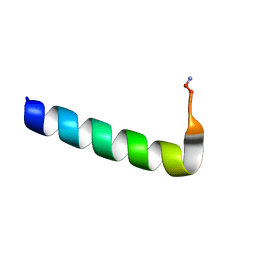 | | Racemic Phenol-Soluble Modulin Alpha 3 Peptide | | Descriptor: | Phenol-soluble modulin PSM-alpha-3 | | Authors: | Yao, Z, Cary, B.P, Bingman, C.A, Wang, C, Kreitler, D.F, Satyshur, K.A, Forest, K.T, Gellman, S.H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Use of a Stereochemical Strategy To Probe the Mechanism of Phenol-Soluble Modulin alpha 3 Toxicity.
J.Am.Chem.Soc., 141, 2019
|
|
7D6F
 
 | | The crystal structure of ARMS-PBM/MAGI2-PDZ4 | | Descriptor: | GLYCEROL, Kinase D-interacting substrate of 220 kDa, Membrane-associated guanylate kinase, ... | | Authors: | Ye, J, Zhang, Y, Zhong, Z, Wang, C. | | Deposit date: | 2020-09-30 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Crystal structure of the PDZ4 domain of MAGI2 in complex with PBM of ARMS reveals a canonical PDZ recognition mode.
Neurochem.Int., 149, 2021
|
|
