1TX7
 
 | | Bovine Trypsin complexed with p-amidinophenylmethylphosphinic acid (AMPA) | | Descriptor: | (4-CARBAMIMIDOYLPHENYL)-METHYL-PHOSPHINIC ACID, CALCIUM ION, Trypsinogen | | Authors: | Cui, J, Marankan, F, Fu, W, Crich, D, Mesecar, A, Johnson, M.E. | | Deposit date: | 2004-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An oxyanion-hole selective serine protease inhibitor in complex with trypsin.
Bioorg.Med.Chem., 10, 2002
|
|
1XH4
 
 | | Crystal Structures of Protein Kinase B Selective Inhibitors in Complex with Protein Kinase A and Mutants | | Descriptor: | N-[4-({4-[5-(DIMETHYLAMINO)-2-HYDROXYBENZOYL]BENZOYL}AMINO)AZEPAN-3-YL]ISONICOTINAMIDE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Breitenlechner, C.B, Friebe, W.-G, Brunet, E, Werner, G, Graul, K, Thomas, U, Kuenkele, K.-P, Schaefer, W, Gassel, M, Bossemeyer, D, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-09-17 | | Release date: | 2005-09-17 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design and crystal structures of protein kinase B-selective inhibitors in complex with protein kinase A and mutants
J.Med.Chem., 48, 2005
|
|
1U39
 
 | | Auto-inhibition Mechanism of X11s/Mints Family Scaffold Proteins Revealed by the Closed Conformation of the Tandem PDZ Domains | | Descriptor: | amyloid beta A4 precursor protein-binding, family A, member 1 | | Authors: | Feng, W, Long, J.-F, Chan, L.-N, He, C, Fu, A, Xia, J, Ip, N.Y, Zhang, M. | | Deposit date: | 2004-07-21 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Autoinhibition of X11/Mint scaffold proteins revealed by the closed conformation of the PDZ tandem
Nat.Struct.Mol.Biol., 12, 2005
|
|
6CC9
 
 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | Descriptor: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | Authors: | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | Deposit date: | 2018-02-06 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
5ZO1
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain (Ig1-Ig3), 2.2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 4, GLYCEROL | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4FU3
 
 | | CID of human RPRD1B | | Descriptor: | CHLORIDE ION, Regulation of nuclear pre-mRNA domain-containing protein 1B, UNKNOWN ATOM OR ION | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Guo, X, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CID of human RPRD1B
TO BE PUBLISHED
|
|
6NGG
 
 | |
6ASG
 
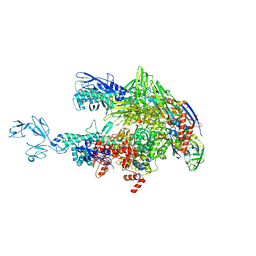 | | Crystal structure of Thermus thermophilus RNA polymerase core enzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, Y, Lin, W, Ying, R, Ebright, R.H. | | Deposit date: | 2017-08-24 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Basis of Transcription Inhibition by Fidaxomicin (Lipiarmycin A3).
Mol. Cell, 70, 2018
|
|
6O0B
 
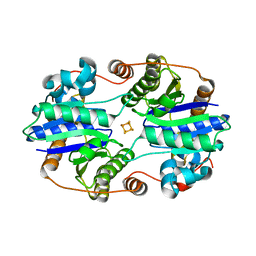 | | Structural and Mechanistic Insights into CO2 Activation by Nitrogenase Iron Protein | | Descriptor: | IRON/SULFUR CLUSTER, Nitrogenase iron protein | | Authors: | Rettberg, L.A, Stiebritz, M.T, Kang, W, Lee, C.C, Ribbe, M.W, Hu, Y. | | Deposit date: | 2019-02-15 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Mechanistic Insights into CO2Activation by Nitrogenase Iron Protein.
Chemistry, 25, 2019
|
|
2O1E
 
 | | Crystal Structure of the Metal-dependent Lipoprotein YcdH from Bacillus subtilis, Northeast Structural Genomics Target SR583 | | Descriptor: | MANGANESE (II) ION, YcdH | | Authors: | Forouhar, F, Zhou, W, Seetharaman, J, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.-C, Janjua, H, Xiao, R, Baran, M.C, Liu, J, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Metal-dependent Lipoprotein YcdH from Bacillus subtilis, Northeast Structural Genomics Target SR583
To be Published
|
|
6NJE
 
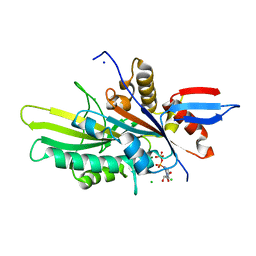 | | Crystal structure of the motor domain of human kinesin family member 22 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Kinesin-like protein KIF22, ... | | Authors: | Walker, B.C, Zhu, H, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Park, H, Cochran, J.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-01-03 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the motor domain of human kinesin family member 22
To Be Published
|
|
6O0W
 
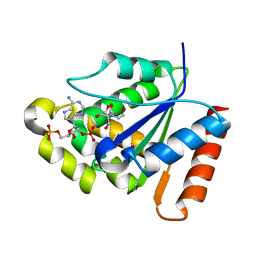 | | Crystal structure of the TIR domain from the grapevine disease resistance protein RUN1 in complex with NADP+ and Bis-Tris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-2'-5'-DIPHOSPHATE, TIR-NB-LRR type resistance protein RUN1 | | Authors: | Horsefield, S, Burdett, H, Zhang, X, Manik, M.K, Shi, Y, Chen, J, Tiancong, Q, Gilley, J, Lai, J, Gu, W, Rank, M, Casey, L, Ericsson, D.J, Foley, G, Hughes, R.O, Bosanac, T, von Itzstein, M, Rathjen, J.P, Nanson, J.D, Boden, M, Dry, I.B, Williams, S.J, Staskawicz, B.J, Coleman, M.P, Ve, T, Dodds, P.N, Kobe, B. | | Deposit date: | 2019-02-17 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | NAD+cleavage activity by animal and plant TIR domains in cell death pathways.
Science, 365, 2019
|
|
1ZKD
 
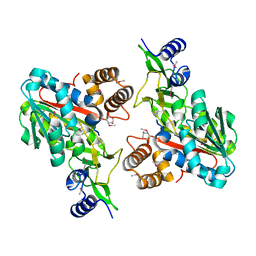 | | X-Ray structure of the putative protein Q6N1P6 from Rhodopseudomonas palustris at the resolution 2.1 A , Northeast Structural Genomics Consortium target RpR58 | | Descriptor: | DUF185 | | Authors: | Kuzin, A.P, Yong, W, Vorobiev, S.M, Acton, T, Ma, L, Xiao, R, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-02 | | Release date: | 2005-05-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray structure of the putative protein Q6N1P6 from Rhodopseudomonas palustris at the resolution 2.1 A , Northeast Structural Genomics Consortium target RpR58
To be Published
|
|
5Z6B
 
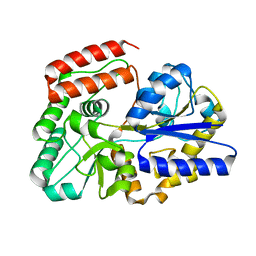 | | Crystal structure of sugar-binding protein YesO in complex with rhamnogalacturonan trisaccharide | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-[beta-D-galactopyranose-(1-4)]alpha-L-rhamnopyranose, Putative ABC transporter substrate-binding protein YesO | | Authors: | Sugiura, H, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2018-01-22 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | Crystal structure of sugar-binding protein YesO in complex with rhamnogalacturonan trisaccharide
To Be Published
|
|
6AYB
 
 | | Naegleria fowleri CYP51-ketoconazole complex | | Descriptor: | 1,2-ETHANEDIOL, 1-acetyl-4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-imidazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazine, CALCIUM ION, ... | | Authors: | Debnath, A, Calvet, C.M, Jennings, G, Zhou, W, Aksenov, A, Luth, M, Abagyan, R, Nes, W.D, McKerrow, J.H, Podust, L.M. | | Deposit date: | 2017-09-08 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | CYP51 is an essential drug target for the treatment of primary amoebic meningoencephalitis (PAM).
PLoS Negl Trop Dis, 11, 2017
|
|
6AZ4
 
 | | RNA hairpin complex with guanosine dinucleotide ligand G(5')ppp(5')G | | Descriptor: | DIGUANOSINE-5'-TRIPHOSPHATE, RNA (32-MER), RNA (5'-R(*CP*AP*GP*CP*AP*GP*CP*AP*G)-3') | | Authors: | Zhang, W, Szostak, J.W. | | Deposit date: | 2017-09-09 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural Rationale for the Enhanced Catalysis of Nonenzymatic RNA Primer Extension by a Downstream Oligonucleotide.
J. Am. Chem. Soc., 140, 2018
|
|
2O6U
 
 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- new crystal form. | | Descriptor: | THIOESTERASE | | Authors: | Chruszcz, M, Koclega, K.D, Evdokimova, E, Cymborowski, M, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-08 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
5ZBR
 
 | | Crystal Structure of Kinesin-3 KIF13B motor domain in AMPPNP form | | Descriptor: | Kinesin family member 13B, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J.Q, Wang, S, Feng, W. | | Deposit date: | 2018-02-12 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Delineation of the Neck Linker of Kinesin-3 for Processive Movement.
J. Mol. Biol., 430, 2018
|
|
1YW6
 
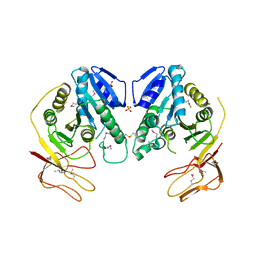 | | Crystal Structure of Succinylglutamate Desuccinylase from Escherichia coli, Northeast Structural Genomics Target ET72. | | Descriptor: | SULFATE ION, Succinylglutamate desuccinylase | | Authors: | Forouhar, F, Yong, W, Kuzin, A.P, Ciano, M, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-17 | | Release date: | 2005-03-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of Succinylglutamate Desuccinylase from Escherichia coli, Northeast Structural Genomics Target ET72.
To be Published
|
|
4HBN
 
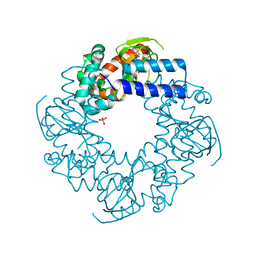 | | Crystal structure of the human HCN4 channel C-terminus carrying the S672R mutation | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, PHOSPHATE ION, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Xu, X, Marni, F, Wu, X, Su, Z, Musayev, F, Shrestha, S, Xie, C, Gao, W, Liu, Q, Zhou, L. | | Deposit date: | 2012-09-28 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Local and Global Interpretations of a Disease-Causing Mutation near the Ligand Entry Path in Hyperpolarization-Activated cAMP-Gated Channel.
Structure, 20, 2012
|
|
2O6B
 
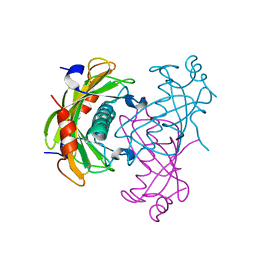 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- new crystal form. | | Descriptor: | THIOESTERASE | | Authors: | Chruszcz, M, Koclega, K.D, Evdokimova, E, Cymborowski, M, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-07 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
4HBK
 
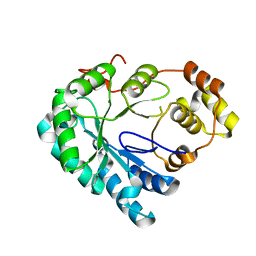 | | Structure of the Aldose Reductase from Schistosoma japonicum | | Descriptor: | Aldo-keto reductase family 1, member B4 (Aldose reductase) | | Authors: | Liu, J, Cheng, J, Zhang, X, Yang, Z, Hu, W, Xu, Y. | | Deposit date: | 2012-09-28 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aldose reductase from Schistosoma japonicum: crystallization and structure-based inhibitor screening for discovering antischistosomal lead compounds.
Parasit Vectors, 6, 2013
|
|
2OFY
 
 | | Crystal structure of putative XRE-family transcriptional regulator from Rhodococcus sp. | | Descriptor: | Putative XRE-family transcriptional regulator | | Authors: | Shumilin, I.A, Skarina, T, Onopriyenko, O, Yim, V, Chruszcz, M, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-01-04 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative XRE-family transcriptional regulator from Rhodococcus sp.
To be Published
|
|
1RQL
 
 | | Crystal Structure of Phosponoacetaldehyde Hydrolase Complexed with Magnesium and the Inhibitor Vinyl Sulfonate | | Descriptor: | MAGNESIUM ION, Phosphonoacetaldehyde Hydrolase, VINYLSULPHONIC ACID | | Authors: | Morais, M.C, Zhang, G, Zhang, W, Olsen, D.B, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-12-05 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic and site-directed mutagenesis
analysis of the mechanism of Schiff-base formation in
phosphonoacetaldehyde hydrolase catalysis
J.Biol.Chem., 279, 2004
|
|
6NVX
 
 | | Crystal structure of penicillin G acylase from Bacillus sp. FJAT-27231 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Blankenfeldt, W. | | Deposit date: | 2019-02-05 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structures and protein engineering of three different penicillin G acylases from Gram-positive bacteria with different thermostability.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
