7ZRV
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, full map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, ... | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7RT5
 
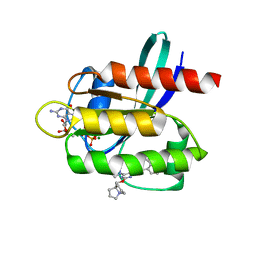 | | Crystal structure of KRAS G12D with compound 36 (4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-7-(8-ethynyl-7-fluoronaphthalen-1-yl)-8-fluoro-2-{[(4s,7as)-tetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidine) bound | | Descriptor: | 4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-7-(8-ethynyl-7-fluoronaphthalen-1-yl)-8-fluoro-2-{[(4s,7as)-tetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
7RT3
 
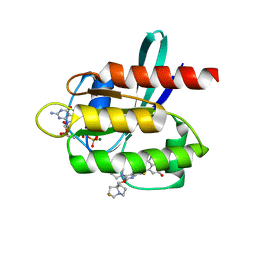 | | Crystal Structure of KRAS G12D with compound 24 (4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S,4S,7aR)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol) bound | | Descriptor: | 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S,4S,7aR)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
7RT1
 
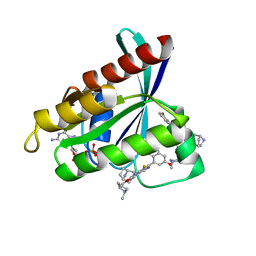 | | Crystal Structure of KRAS G12D with compound 15 (4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol) bound | | Descriptor: | 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
7RT2
 
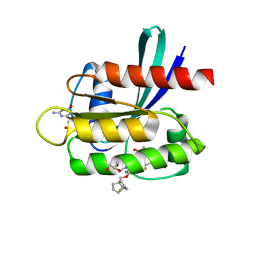 | | Crystal Structure of KRAS G12D with compound 25 (4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol) bound | | Descriptor: | 1,2-ETHANEDIOL, 4-(4-[(1R,5S)-3,8-diazabicyclo[3.2.1]octan-3-yl]-8-fluoro-2-{[(2R,4R,7aS)-2-fluorotetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}pyrido[4,3-d]pyrimidin-7-yl)naphthalen-2-ol, GLYCEROL, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
7RT4
 
 | | KRAS G12D in complex with Compound 5B (7-(8-chloronaphthalen-1-yl)-8-fluoro-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}-4-(piperazin-1-yl)pyrido[4,3-d]pyrimidine) | | Descriptor: | 7-(8-chloronaphthalen-1-yl)-8-fluoro-2-{[(2S)-1-methylpyrrolidin-2-yl]methoxy}-4-(piperazin-1-yl)pyrido[4,3-d]pyrimidine, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gunn, R.J, Thomas, N.C, Xiaolun, W, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-08-12 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS G12D Inhibitor.
J.Med.Chem., 65, 2022
|
|
7SCP
 
 | | The crystal structure of ScoE in complex with intermediate | | Descriptor: | (3R)-3-(oxaloamino)butanoic acid, 1,2-ETHANEDIOL, FE (II) ION, ... | | Authors: | Cha, L, Chen, J, Zhou, J, Chang, W. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Deciphering the Reaction Pathway of Mononuclear Iron Enzyme-Catalyzed N-C Triple Bond Formation in Isocyanide Lipopeptide and Polyketide Biosynthesis
Acs Catalysis, 12, 2022
|
|
8ESM
 
 | | Human triacylglycerol synthesizing enzyme DGAT1 in complex with T863 inhibitor | | Descriptor: | Diacylglycerol O-acyltransferase 1, {(1r,4r)-4-[4-(4-amino-7,7-dimethyl-7H-pyrimido[4,5-b][1,4]oxazin-6-yl)phenyl]cyclohexyl}acetic acid | | Authors: | Sui, X, Kun, W, Walther, T, Farese, R, Liao, M. | | Deposit date: | 2022-10-14 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of action for small-molecule inhibitors of triacylglycerol synthesis.
Nat Commun, 14, 2023
|
|
8ETM
 
 | | Human triacylglycerol synthesizing enzyme DGAT1 in complex with DGAT1IN1 inhibitor | | Descriptor: | Diacylglycerol O-acyltransferase 1, [(1S,4r)-4-{4-[(4S)-2-({[4-(trifluoromethoxy)phenyl]methyl}carbamoyl)imidazo[1,2-a]pyridin-6-yl]phenyl}cyclohexyl]acetic acid | | Authors: | Sui, X, Kun, W, Walther, T, Farese, R, Liao, M. | | Deposit date: | 2022-10-17 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of action for small-molecule inhibitors of triacylglycerol synthesis.
Nat Commun, 14, 2023
|
|
8EPN
 
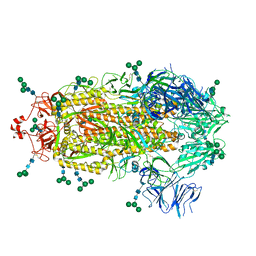 | |
8EPQ
 
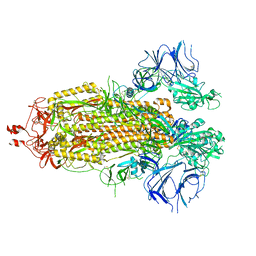 | | Cryo-EM structure of SARS-CoV-2 Spike trimer S2D14 with two RBDs exposed | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Williams, J.A, Harshbarger, W. | | Deposit date: | 2022-10-06 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and computational design of a SARS-CoV-2 spike antigen with improved expression and immunogenicity.
Sci Adv, 9, 2023
|
|
8EPP
 
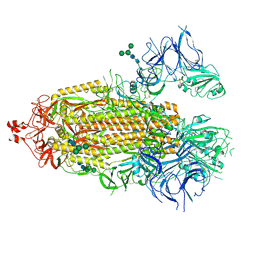 | |
8FD5
 
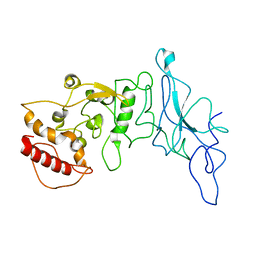 | | Nucleocapsid monomer structure from SARS-CoV-2 | | Descriptor: | Nucleoprotein | | Authors: | Casasanta, M, Jonaid, G.M, Kaylor, L, Luqiu, W, DiCecco, L, Solares, M, Berry, S, Kelly, D.F. | | Deposit date: | 2022-12-02 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (4.57 Å) | | Cite: | Structural Insights of the SARS-CoV-2 Nucleocapsid Protein: Implications for the Inner-workings of Rapid Antigen Tests.
Microsc Microanal, 29, 2023
|
|
8GMN
 
 | | Crystal structure of human C1s in complex with inhibitor | | Descriptor: | Complement C1s subcomponent, SULFATE ION, [4-(1-aminophthalazin-6-yl)piperazin-1-yl](2-methylphenyl)methanone | | Authors: | Dougan, D.R, Lane, W. | | Deposit date: | 2023-03-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of a Novel Series of Potent, Selective, Orally Available, and Brain-Penetrable C1s Inhibitors for Modulation of the Complement Pathway.
J.Med.Chem., 66, 2023
|
|
8G5C
 
 | |
8G6U
 
 | | Cryo-EM structure of T/F100 SOSIP.664 HIV-1 Env trimer with LMHS mutations in complex with 8ANC195 and 10-1074 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CRF-1_AE T/F100 HIV-1 gp41, ... | | Authors: | Chen, Y, Zhou, F, Huang, R, Tolbert, W, Pazgier, M. | | Deposit date: | 2023-02-16 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir.
Nat Commun, 14, 2023
|
|
8FDT
 
 | | Engineered human dynein motor domain in the microtubule-unbound state with LIS1 complex in the buffer containing ATP-Vi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1,Serine--tRNA ligase, ... | | Authors: | Ton, W, Wang, Y, Chai, P. | | Deposit date: | 2022-12-04 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Microtubule-binding-induced allostery triggers LIS1 dissociation from dynein prior to cargo transport.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FDU
 
 | | Engineered human dynein motor domain in the microtubule-unbound state with LIS1 complex in the buffer containing ATP-Vi (local refined on AAA3-AAA5 and LIS1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1,Serine--tRNA ligase, Platelet-activating factor acetylhydrolase IB subunit beta,Platelet-activating factor acetylhydrolase IB subunit beta,human LIS1 protein with a SNAP tag | | Authors: | Ton, W, Wang, Y, Chai, P. | | Deposit date: | 2022-12-04 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Microtubule-binding-induced allostery triggers LIS1 dissociation from dynein prior to cargo transport.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FCY
 
 | | Engineered human dynein motor domain in microtubule-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1,Serine--tRNA ligase | | Authors: | Ton, W, Wang, Y, Chai, P. | | Deposit date: | 2022-12-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Microtubule-binding-induced allostery triggers LIS1 dissociation from dynein prior to cargo transport.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FD6
 
 | | Engineered human dynein motor domain in the microtubule-unbound state in the buffer containing ATP-Vi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1,Serine--tRNA ligase, ... | | Authors: | Ton, W, Wang, Y, Chai, P. | | Deposit date: | 2022-12-02 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Microtubule-binding-induced allostery triggers LIS1 dissociation from dynein prior to cargo transport.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2WFA
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Beryllium trifluoride, in an open conformation. | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Near Attack Conformers Dominate Beta-Phosphoglucomutase Complexes Where Geometry and Charge Distribution Reflect Those of Substrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5IT5
 
 | | Thermus thermophilus PilB core ATPase region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP binding motif-containing protein PilF, MAGNESIUM ION, ... | | Authors: | Mancl, J, Robinson, H, Black, W, Yang, Z, Schubot, F. | | Deposit date: | 2016-03-16 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Crystal Structure of a Type IV Pilus Assembly ATPase: Insights into the Molecular Mechanism of PilB from Thermus thermophilus.
Structure, 24, 2016
|
|
5I4R
 
 | | Contact-dependent inhibition system from Escherichia coli NC101 - ternary CdiA/CdiI/EF-Tu complex (trypsin-modified) | | Descriptor: | Contact-dependent inhibitor A, Contact-dependent inhibitor I, Elongation factor Tu, ... | | Authors: | Michalska, K, Stols, L, Eschenfeldt, W, Hayes, C.S, Goulding, C.W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI) | | Deposit date: | 2016-02-12 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of a novel antibacterial toxin that exploits elongation factor Tu to cleave specific transfer RNAs.
Nucleic Acids Res., 45, 2017
|
|
2X47
 
 | | Crystal structure of human MACROD1 | | Descriptor: | MACRO DOMAIN-CONTAINING PROTEIN 1, SULFATE ION | | Authors: | Vollmar, M, Phillips, C, Mehrotra, P.V, Ahel, I, Krojer, T, Yue, W, Ugochukwu, E, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Gileadi, O. | | Deposit date: | 2010-01-28 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of Macro Domain Proteins as Novel O-Acetyl-Adp-Ribose Deacetylases.
J.Biol.Chem., 286, 2011
|
|
2WNL
 
 | | CRYSTAL STRUCTURE OF APLYSIA ACHBP IN COMPLEX WITH ANABASEINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,4,5,6-tetrahydro-2,3'-bipyridine, 5-amino-1-pyridin-3-ylpentan-1-one, ... | | Authors: | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | Deposit date: | 2009-07-09 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal alpha7 nicotinic acetylcholine receptor.
EMBO J., 28, 2009
|
|
