3F5K
 
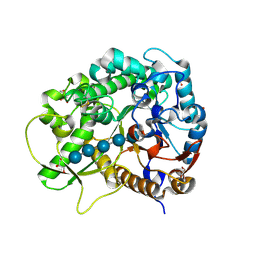 | | Semi-active E176Q mutant of rice BGlu1, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-glucosidase, GLYCEROL, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2008-11-03 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of oligosaccharide binding by rice BGlu1 beta-glucosidase
J.Struct.Biol., 173, 2011
|
|
3HXI
 
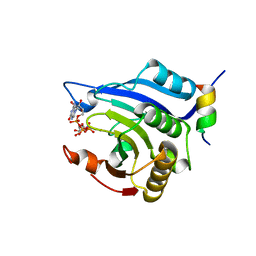 | | Crystal structure of Schistosome eIF4E complexed with m7GpppG and 4E-BP | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Eukaryotic Translation Initiation 4E, Eukaryotic translation initiation factor 4E-binding protein 1 | | Authors: | Liu, W, Zhao, R, Jones, D.N.M, Davis, R.E. | | Deposit date: | 2009-06-20 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into parasite EIF4E binding specificity for m7G and m2,2,7G mRNA cap.
J.Biol.Chem., 284, 2009
|
|
2K20
 
 | |
1ONP
 
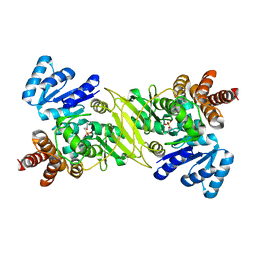 | | IspC complex with Mn2+ and fosmidomycin | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MANGANESE (II) ION | | Authors: | Steinbacher, S, Kaiser, J, Eisenreich, W, Huber, R, Bacher, A, Rohdich, F. | | Deposit date: | 2003-02-28 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of fosmidomycin action revealed by the complex with 2-C-methyl-D-erythritol
4-phosphate synthase (IspC). Implications for the catalytic mechanism and anti-malaria
drug development.
J.BIOL.CHEM., 278, 2003
|
|
3EZ2
 
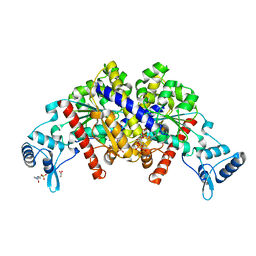 | | Partition protein-ADP complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Schumacher, M.A, Dunham, T.D, Xu, W, Funnell, B. | | Deposit date: | 2008-10-22 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA.
Embo J., 28, 2009
|
|
3F0C
 
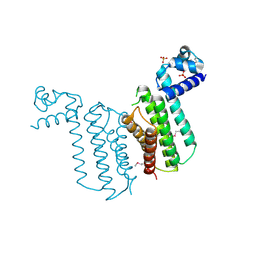 | | Crystal structure of transcriptional regulator from Cytophaga hutchinsonii ATCC 33406 | | Descriptor: | SULFATE ION, Transcriptional regulator | | Authors: | Nocek, B, Maltseva, N, Tan, K, Abdullah, J, Eschenfeldt, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal structure of transcriptional regulator from Cytophaga hutchinsonii ATCC 33406
To be Published
|
|
3I4A
 
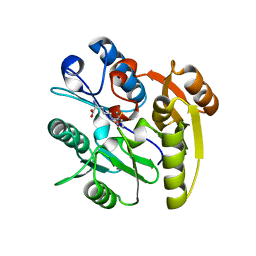 | | Crystal structure of dimethylarginine dimethylaminohydrolase-1 (DDAH-1) in complex with N5-(1-iminopropyl)-L-ornithine | | Descriptor: | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1, N5-(1-iminopropyl)-L-ornithine | | Authors: | Monzingo, A.F, Wang, Y, Hu, S, Schaller, T.H, Fast, W, Robertus, J.D. | | Deposit date: | 2009-07-01 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Developing dual and specific inhibitors of dimethylarginine dimethylaminohydrolase-1 and nitric oxide synthase: toward a targeted polypharmacology to control nitric oxide.
Biochemistry, 48, 2009
|
|
3I5W
 
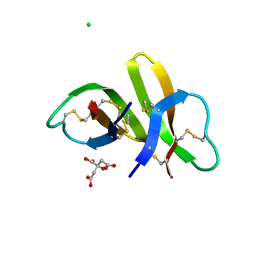 | | Crystal structure of human alpha-defensin 5 (mutant R13H) | | Descriptor: | CHLORIDE ION, CITRATE ANION, Defensin-5 | | Authors: | Pazgier, M, Lu, W. | | Deposit date: | 2009-07-06 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Selective arginines are important for the antibacterial activity and host cell interaction of human alpha-defensin 5
Febs Lett., 583, 2009
|
|
7ZCH
 
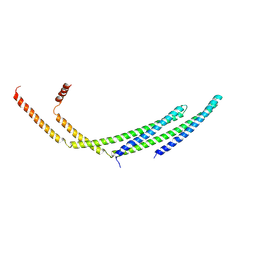 | | CHMP2A-CHMP3 heterodimer (410 Angstrom diameter) | | Descriptor: | Charged multivesicular body protein 2a, Charged multivesicular body protein 3 | | Authors: | Azad, K, Desfosses, A, Effantin, G, Schoehn, G, Weissenhorn, W. | | Deposit date: | 2022-03-28 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of CHMP2A-CHMP3 ESCRT-III polymer assembly and membrane cleavage.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3IZJ
 
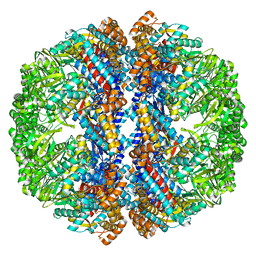 | | Mm-cpn rls with ATP and AlFx | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
1ORV
 
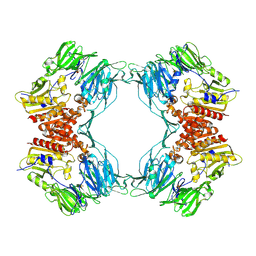 | | Crystal Structure of Porcine Dipeptidyl Peptidase IV (CD26) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Engel, M, Hoffmann, T, Wagner, L, Wermann, M, Heiser, U, Kiefersauer, R, Huber, R, Bode, W, Demuth, H.U, Brandstetter, H. | | Deposit date: | 2003-03-16 | | Release date: | 2003-05-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Dipeptidyl Peptidase IV (CD26) Reveals its Functional Regulation and Enzymatic Mechanism
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3J0G
 
 | |
7ZCG
 
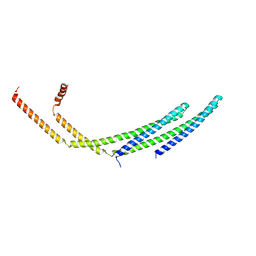 | | CHMP2A-CHMP3 heterodimer (430 Angstrom diameter) | | Descriptor: | Charged multivesicular body protein 2a, Charged multivesicular body protein 3 | | Authors: | Azad, K, Desfosses, A, Effantin, G, Schoehn, G, Weissenhorn, W. | | Deposit date: | 2022-03-28 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of CHMP2A-CHMP3 ESCRT-III polymer assembly and membrane cleavage.
Nat.Struct.Mol.Biol., 30, 2023
|
|
3F5X
 
 | | CDK-2-Cyclin complex with indazole inhibitor 9 bound at its active site | | Descriptor: | 4-{3-[7-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-11-04 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2JZ1
 
 | | DSX_long | | Descriptor: | Protein doublesex | | Authors: | Yang, Y, Zhang, W, Bayrer, J.R, Weiss, M.A. | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Doublesex and the Regulation of Sexual Dimorphism in Drosophila melanogaster: STRUCTURE, FUNCTION, AND MUTAGENESIS OF A FEMALE-SPECIFIC DOMAIN.
J.Biol.Chem., 283, 2008
|
|
1P0N
 
 | | IPP:DMAPP isomerase type II, FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, Isopentenyl-diphosphate delta-isomerase | | Authors: | Steinbacher, S, Kaiser, J, Gerhardt, S, Eisenreich, W, Huber, R, Bacher, A, Rohdich, F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Type II Isopentenyl Diphosphate:Dimethylallyl Diphosphate Isomerase from Bacillus subtilis
J.Mol.Biol., 329, 2003
|
|
1P1D
 
 | |
1P49
 
 | | Structure of Human Placental Estrone/DHEA Sulfatase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Hernandez-Guzman, F.G, Higashiyama, T, Pangborn, W, Osawa, Y, Ghosh, D. | | Deposit date: | 2003-04-21 | | Release date: | 2003-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Human Estrone Sulfatase Suggests Functional Roles of Membrane Association
J.Biol.Chem., 278, 2003
|
|
7W1N
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KRP | | Descriptor: | 1,2-ETHANEDIOL, BICINE, Leaf-branch compost cutinase | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W44
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_RIP | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Leaf-branch compost cutinase | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7WI6
 
 | | Cryo-EM structure of LY341495/NAM-bound mGlu3 | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
7W45
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KIP | | Descriptor: | CALCIUM ION, Leaf-branch compost cutinase, SODIUM ION | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7WIH
 
 | | Cryo-EM structure of LY2794193-bound mGlu3 | | Descriptor: | (1S,2S,4S,5R,6S)-2-amino-4-[(3-methoxybenzene-1-carbonyl)amino]bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
3ISU
 
 | | Crystal structure of the RGC domain of IQGAP3 | | Descriptor: | Ras GTPase-activating-like protein IQGAP3 | | Authors: | Nedyalkova, L, Tempel, W, Tong, Y, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of the RGC domain of IQGAP3
to be published
|
|
7WI8
 
 | | Cryo-EM structure of inactive mGlu3 bound to LY341495 | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 3 | | Authors: | Fang, W, Yang, F, Xu, C.J, Ling, S.L, Lin, L, Zhou, Y.X, Sun, W.J, Wang, X.M, Liu, P, Rondard, P, Pan, S, Pin, J.P, Tian, C.L, Liu, J.F. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structural basis of the activation of metabotropic glutamate receptor 3.
Cell Res., 32, 2022
|
|
