1LI1
 
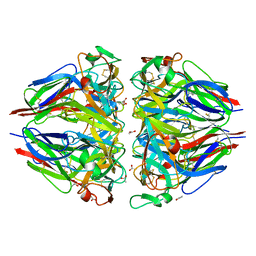 | | The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link | | Descriptor: | ACETATE ION, Collagen alpha 1(IV), Collagen alpha 2(IV) | | Authors: | Than, M.E, Henrich, S, Huber, R, Ries, A, Mann, K, Kuhn, K, Timpl, R, Bourenkov, G.P, Bartunik, H.D, Bode, W. | | Deposit date: | 2002-04-17 | | Release date: | 2002-05-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1C52
 
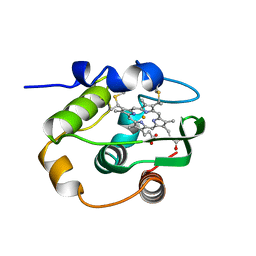 | | THERMUS THERMOPHILUS CYTOCHROME-C552: A NEW HIGHLY THERMOSTABLE CYTOCHROME-C STRUCTURE OBTAINED BY MAD PHASING | | Descriptor: | CYTOCHROME-C552, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Than, M.E, Hof, P, Huber, R, Bourenkov, G.P, Bartunik, H.D, Buse, G, Soulimane, T. | | Deposit date: | 1997-06-23 | | Release date: | 1998-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Thermus thermophilus cytochrome-c552: A new highly thermostable cytochrome-c structure obtained by MAD phasing.
J.Mol.Biol., 271, 1997
|
|
1S4V
 
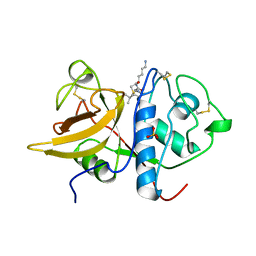 | | The 2.0 A crystal structure of the KDEL-tailed cysteine endopeptidase functioning in programmed cell death of Ricinus communis endosperm | | Descriptor: | DVA-LEU-LYS-0QE peptide, SULFATE ION, cysteine endopeptidase | | Authors: | Than, M.E, Helm, M, Simpson, D.J, Lottspeich, F, Huber, R, Gietl, C. | | Deposit date: | 2004-01-19 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A crystal structure and substrate specificity of the KDEL-tailed cysteine endopeptidase functioning in programmed cell death of Ricinus communis endosperm.
J.Mol.Biol., 336, 2004
|
|
4LXC
 
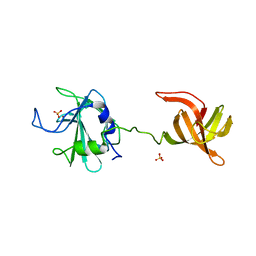 | | The antimicrobial peptidase lysostaphin from Staphylococcus simulans | | Descriptor: | Lysostaphin, SULFATE ION, ZINC ION | | Authors: | Sabala, I, Jagielska, E, Bardelang, P.T, Czapinska, H, Dahms, S.O, Sharpe, J.A, James, R, Than, M.E, Thomas, N.R, Bochtler, M. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the antimicrobial peptidase lysostaphin from Staphylococcus simulans.
Febs J., 281, 2014
|
|
6EQW
 
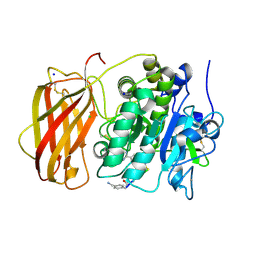 | |
6EQV
 
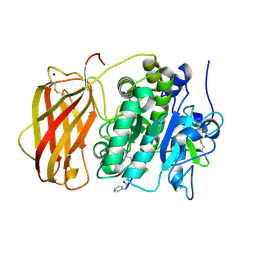 | |
1EHK
 
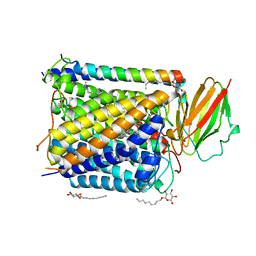 | | CRYSTAL STRUCTURE OF THE ABERRANT BA3-CYTOCHROME-C OXIDASE FROM THERMUS THERMOPHILUS | | Descriptor: | BA3-TYPE CYTOCHROME-C OXIDASE, COPPER (II) ION, DINUCLEAR COPPER ION, ... | | Authors: | Soulimane, T, Buse, G, Bourenkov, G.P, Bartunik, H.D, Huber, R, Than, M.E. | | Deposit date: | 2000-02-21 | | Release date: | 2001-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of the aberrant ba(3)-cytochrome c oxidase from thermus thermophilus.
EMBO J., 19, 2000
|
|
5MIM
 
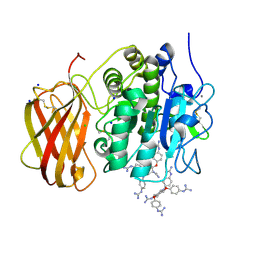 | | Xray structure of human furin bound with the 2,5-dideoxystreptamine derived small molecule inhibitor 1n | | Descriptor: | 1-[(1~{R},2~{R},4~{S},5~{S})-2,4-bis(4-carbamimidamidophenoxy)-5-[(4-carbamimidamidophenyl)amino]cyclohexyl]guanidine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Guan-Sheng, J, Than, M.E. | | Deposit date: | 2016-11-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Studies Revealed Active Site Distortions of Human Furin by a Small Molecule Inhibitor.
ACS Chem. Biol., 12, 2017
|
|
5JXJ
 
 | | Structure of the proprotein convertase furin complexed to meta-guanidinomethyl-Phac-RVR-Amba in presence of EDTA | | Descriptor: | 2UC-ARG-VAL-ARG-00S, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JXI
 
 | | Structure of the unliganded form of the proprotein convertase furin in presence of EDTA. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Furin, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JXG
 
 | | Structure of the unliganded form of the proprotein convertase furin. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Furin, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JMR
 
 | | X-ray structure of the furin inhibitory antibody Nb14 | | Descriptor: | CHLORIDE ION, camelid VHH fragment | | Authors: | Dahms, S.O, Than, M.E. | | Deposit date: | 2016-04-29 | | Release date: | 2016-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The structure of a furin-antibody complex explains non-competitive inhibition by steric exclusion of substrate conformers.
Sci Rep, 6, 2016
|
|
5JMO
 
 | |
5JXH
 
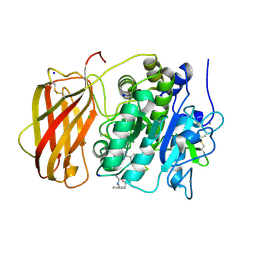 | | Structure the proprotein convertase furin in complex with meta-guanidinomethyl-Phac-RVR-Amba at 2.0 Angstrom resolution. | | Descriptor: | 2UC-ARG-VAL-ARG-00S, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6EQX
 
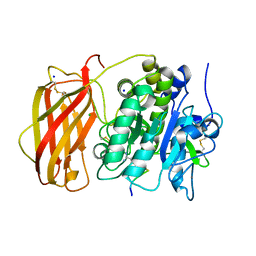 | |
3QO4
 
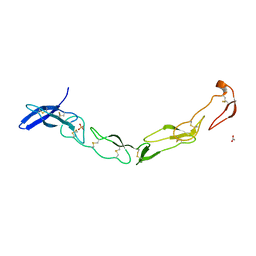 | | The Crystal Structure of Death Receptor 6 | | Descriptor: | ACETATE ION, SULFATE ION, Tumor necrosis factor receptor superfamily member 21 | | Authors: | Kuester, M, Kemmerzehl, S, Dahms, S.O, Roeser, D, Than, M.E. | | Deposit date: | 2011-02-09 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of death receptor 6 (DR6): a potential receptor of the amyloid precursor protein (APP).
J.Mol.Biol., 409, 2011
|
|
4QPB
 
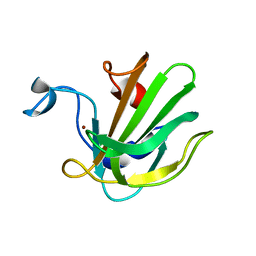 | | Catalytic domain of the antimicrobial peptidase lysostaphin from Staphylococcus simulans crystallized in the absence of phosphate | | Descriptor: | 1,2-ETHANEDIOL, Lysostaphin, ZINC ION | | Authors: | Sabala, I, Jagielska, E, Bardelang, P.T, Czapinska, H, Dahms, S.O, Sharpe, J.A, James, R, Than, M.E, Thomas, N.R, Bochtler, M. | | Deposit date: | 2014-06-22 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the antimicrobial peptidase lysostaphin from Staphylococcus simulans.
Febs J., 281, 2014
|
|
4RD9
 
 | |
4RDA
 
 | | X-RAY STRUCTURE OF THE AMYLOID PRECURSOR PROTEIN-LIKE PROTEIN 1 (APLP1) E2 DOMAIN IN COMPLEX WITH A HEPARIN DODECASACCHARIDE | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, Amyloid-like protein 1, ... | | Authors: | Dahms, S.O, Than, M.E. | | Deposit date: | 2014-09-18 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Interaction of the amyloid precursor protein-like protein 1 (APLP1) E2 domain with heparan sulfate involves two distinct binding modes.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RYD
 
 | |
3KTM
 
 | | Structure of the Heparin-induced E1-Dimer of the Amyloid Precursor Protein (APP) | | Descriptor: | (3R)-butane-1,3-diol, ACETATE ION, Amyloid beta A4 protein, ... | | Authors: | Dahms, S.O, Hoefgen, S, Roeser, D, Schlott, B, Guhrs, K.H, Than, M.E. | | Deposit date: | 2009-11-25 | | Release date: | 2010-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and biochemical analysis of the heparin-induced E1 dimer of the amyloid precursor protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4OMC
 
 | |
4OMD
 
 | |
4PWQ
 
 | |
4QP5
 
 | | Catalytic domain of the antimicrobial peptidase lysostaphin from Staphylococcus simulans crystallized in the presence of phosphate | | Descriptor: | GLYCEROL, Lysostaphin, PHOSPHATE ION, ... | | Authors: | Sabala, I, Jagielska, E, Bardelang, P.T, Czapinska, H, Dahms, S.O, Sharpe, J.A, James, R, Than, M.E, Thomas, N.R, Bochtler, M. | | Deposit date: | 2014-06-22 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of the antimicrobial peptidase lysostaphin from Staphylococcus simulans.
Febs J., 281, 2014
|
|
