2DO3
 
 | | Solution structure of the third KOW motif of transcription elongation factor SPT5 | | 分子名称: | Transcription elongation factor SPT5 | | 著者 | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-04-27 | | 公開日 | 2006-10-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the third KOW motif of transcription elongation factor SPT5
To be Published
|
|
2DO4
 
 | | Solution structure of the RNA binding domain of squamous cell carcinoma antigen recognized by T cells 3 | | 分子名称: | Squamous cell carcinoma antigen recognized by T-cells 3 | | 著者 | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2006-04-27 | | 公開日 | 2007-04-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the RNA binding domain of squamous cell carcinoma antigen recognized by T cells 3
To be Published
|
|
2YQF
 
 | | Solution structure of the death domain of Ankyrin-1 | | 分子名称: | Ankyrin-1 | | 著者 | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-30 | | 公開日 | 2008-04-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the death domain of Ankyrin-1
To be Published
|
|
2YQE
 
 | | Solution structure of the ARID domain of JARID1D protein | | 分子名称: | Jumonji/ARID domain-containing protein 1D | | 著者 | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-30 | | 公開日 | 2008-04-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the ARID domain of JARID1D protein
To be Published
|
|
2YRK
 
 | | Solution structure of the zf-C2H2 domain in zinc finger homeodomain 4 | | 分子名称: | ZINC ION, Zinc finger homeobox protein 4 | | 著者 | Tanabe, W, Suzuki, S, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-04-02 | | 公開日 | 2007-10-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the zf-C2H2 domain in zinc finger homeodomain 4
To be Published
|
|
5XIV
 
 | | Beta-Ginkgotides: Hyperdisulfide-constrained peptides from Ginkgo biloba | | 分子名称: | beta-ginkgotide, beta-gB1 | | 著者 | Wong, K.H, Tan, W.L, Xiao, T, Tam, J.P. | | 登録日 | 2017-04-27 | | 公開日 | 2017-05-31 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | beta-Ginkgotides: Hyperdisulfide-constrained peptides from Ginkgo biloba.
Sci Rep, 7, 2017
|
|
4MOD
 
 | | Structure of the MERS-CoV fusion core | | 分子名称: | HR1 of S protein, LINKER, HR2 of S protein | | 著者 | Gao, J, Lu, G, Qi, J, Li, Y, Wu, Y, Deng, Y, Geng, H, Xiao, H, Tan, W, Yan, J, Gao, G.F. | | 登録日 | 2013-09-12 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of Middle East respiratory syndrome coronavirus.
J.Virol., 87, 2013
|
|
5B7C
 
 | |
5BT2
 
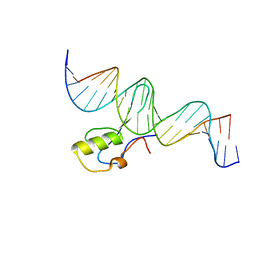 | | MeCP2 MBD domain (A140V) in complex with methylated DNA | | 分子名称: | DNA (5'-D(*AP*TP*AP*GP*AP*AP*GP*AP*AP*TP*TP*CP*(5CM)P*GP*TP*TP*CP*CP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*GP*AP*AP*(5CM)P*GP*GP*AP*AP*TP*TP*CP*TP*TP*CP*TP*A)-3'), Methyl-CpG-binding protein 2 | | 著者 | Ho, K.L, Chia, J.Y, Tan, W.S, Ng, C.L, Hu, N.J, Foo, H.L. | | 登録日 | 2015-06-02 | | 公開日 | 2016-08-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A/T Run Geometry of B-form DNA Is Independent of Bound Methyl-CpG Binding Domain, Cytosine Methylation and Flanking Sequence.
Sci Rep, 6, 2016
|
|
4M1C
 
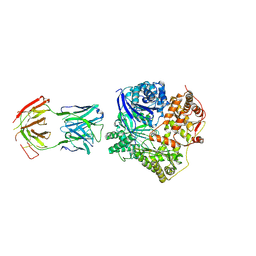 | | Crystal Structure Analysis of Fab-Bound Human Insulin Degrading Enzyme (IDE) in Complex with Amyloid-Beta (1-40) | | 分子名称: | Amyloid beta A4 protein, Fab-bound IDE, heavy chain, ... | | 著者 | McCord, L.M, Liang, W, Farcasanu, M, Scherpelz, K, Meredith, S.C, Koide, S, Tang, W.J. | | 登録日 | 2013-08-02 | | 公開日 | 2014-08-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.5007 Å) | | 主引用文献 | Crystal Structure Analysis of Fab-Bound Human Insulin Degrading Enzyme (IDE) in Complex with Amyloid-Beta (1-40)
To be Published
|
|
7RZI
 
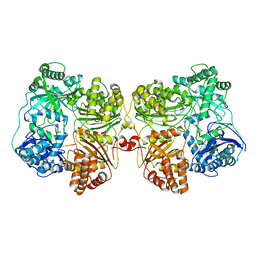 | | Insulin Degrading Enzyme pC/pC | | 分子名称: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | 著者 | Mancl, J.M, Liang, W.G, Tang, W.J. | | 登録日 | 2021-08-27 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZE
 
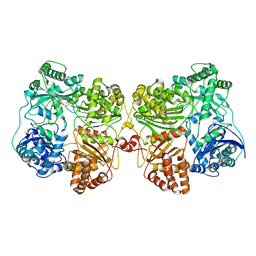 | | Insulin Degrading Enzyme pO/pC | | 分子名称: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | 著者 | Mancl, J.M, Liang, W.G, Tang, W.J. | | 登録日 | 2021-08-27 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZH
 
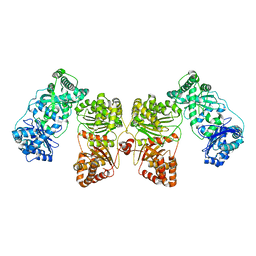 | | Insulin Degrading Enzyme O/O | | 分子名称: | Cysteine-free Insulin-degrading enzyme | | 著者 | Mancl, J.M, Liang, W.G, Tang, W.J. | | 登録日 | 2021-08-27 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZF
 
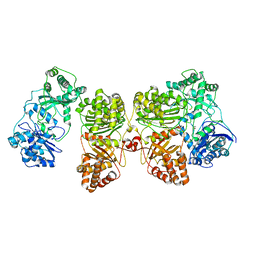 | | Insulin Degrading Enzyme O/pC | | 分子名称: | Cysteine-free Insulin-degrading enzyme, Insulin A chain, Insulin B chain | | 著者 | Mancl, J.M, Liang, W.G, Tang, W.J. | | 登録日 | 2021-08-27 | | 公開日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
7RZG
 
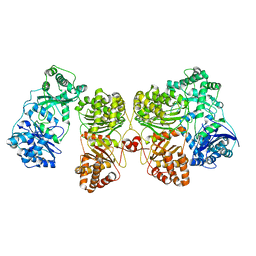 | | Insulin Degrading Enzyme O/pO | | 分子名称: | Cysteine-free Insulin-degrading enzyme | | 著者 | Mancl, J.M, Liang, W.G, Tang, W.J. | | 登録日 | 2021-08-27 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Ensemble cryoEM reveals a substrate-induced shift in the conformational dynamics of human insulin degrading enzyme
To be published
|
|
5NGN
 
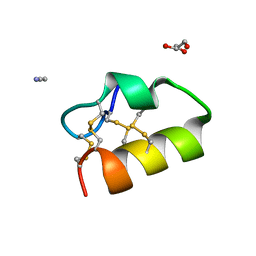 | | Lybatide 2, a cystine-rich peptide from Lycium barbarum | | 分子名称: | ACETONITRILE, GLYCEROL, TETRAETHYLENE GLYCOL, ... | | 著者 | Lei, J, Tan, W.L, Sakai, N, Hilgenfeld, R. | | 登録日 | 2017-03-18 | | 公開日 | 2017-07-26 | | 最終更新日 | 2018-04-11 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Lybatides from Lycium barbarum Contain An Unusual Cystine-stapled Helical Peptide Scaffold.
Sci Rep, 7, 2017
|
|
4DWK
 
 | | Structure of cystein free insulin degrading enzyme with compound bdm41671 ((s)-2-{2-[carboxymethyl-(3-phenyl-propyl)-amino]-acetylamino}-3-(1h-imidazol-4-yl)-propionic acid methyl ester) | | 分子名称: | Insulin-degrading enzyme, ZINC ION, methyl N-(carboxymethyl)-N-(3-phenylpropyl)glycyl-L-histidinate | | 著者 | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | 登録日 | 2012-02-24 | | 公開日 | 2013-04-03 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, substrate-dependent modulators of insulin-degrading enzyme in amyloid-beta hydrolysis.
Eur.J.Med.Chem., 79, 2014
|
|
4LWZ
 
 | | Crystal structure of Myo5b globular tail domain in complex with inactive Rab11a | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-11A, ... | | 著者 | Pylypenko, O, Attanda, W, Gauquelin, C, Houdusse, A. | | 登録日 | 2013-07-29 | | 公開日 | 2013-11-20 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural basis of myosin V Rab GTPase-dependent cargo recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8H0J
 
 | | Annexin A5 mutant | | 分子名称: | Annexin A5, CALCIUM ION | | 著者 | Hua, Z.C, Tang, W. | | 登録日 | 2022-09-29 | | 公開日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | structure dissection of the membrane aggregation mechanism induced by Annexin A5 mutation
To Be Published
|
|
5GRJ
 
 | | Crystal structure of human PD-L1 with monoclonal antibody avelumab | | 分子名称: | Programmed cell death 1 ligand 1, avelumab H chain, avelumab L chain | | 著者 | Liu, K, Tan, S, Chai, Y, Chen, D, Song, H, Zhang, C.W.-H, Shi, Y, Liu, J, Tan, W, Lyu, J, Gao, S, Yan, J, Qi, J, Gao, G.F. | | 登録日 | 2016-08-11 | | 公開日 | 2016-11-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.206 Å) | | 主引用文献 | Structural basis of anti-PD-L1 monoclonal antibody avelumab for tumor therapy.
Cell Res., 27, 2017
|
|
6H2B
 
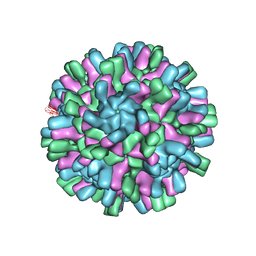 | | Structure of the Macrobrachium rosenbergii Nodavirus | | 分子名称: | CALCIUM ION, Capsid protein | | 著者 | Ho, K.H, Gabrielsen, M, Beh, P.L, Kueh, C.L, Thong, Q.X, Streetley, J, Tan, W.S, Bhella, D. | | 登録日 | 2018-07-13 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | Structure of the Macrobrachium rosenbergii nodavirus: A new genus within the Nodaviridae?
PLoS Biol., 16, 2018
|
|
6B70
 
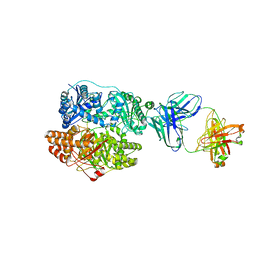 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain and insulin | | 分子名称: | FAB H11-E heavy chain, FAB H11-E light chain, Insulin, ... | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-03 | | 公開日 | 2017-12-27 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF8
 
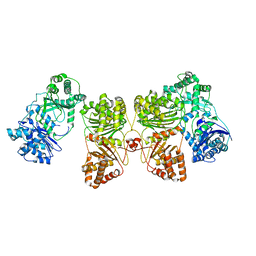 | | Cryo-EM structure of human insulin degrading enzyme in complex with insulin | | 分子名称: | Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF7
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | 分子名称: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2018-02-07 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF9
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | 分子名称: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-26 | | 公開日 | 2018-02-07 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (7.2 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
