6J6U
 
 | | Rat PTPRZ D1-D2 domain | | Descriptor: | Receptor-type tyrosine-protein phosphatase zeta | | Authors: | Sugawara, H. | | Deposit date: | 2019-01-15 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | A head-to-toe dimerization has physiological relevance for ligand-induced inactivation of protein tyrosine receptor type Z.
J.Biol.Chem., 294, 2019
|
|
5Y9L
 
 | | Human kallikrein 7 in complex with 1,3,6-trisubstituted 1,4-diazepane-7-one | | Descriptor: | 3-[2-[(3Z,6R)-6-[(5-chloranyl-2-methoxy-phenyl)methyl]-3-(dimethylhydrazinylidene)-7-oxidanylidene-1,4-diazepan-1-yl]ethanoylamino]benzoic acid, CHLORIDE ION, Kallikrein-7 | | Authors: | Sugawara, H. | | Deposit date: | 2017-08-25 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery and structure-activity relationship study of 1,3,6-trisubstituted 1,4-diazepane-7-ones as novel human kallikrein 7 inhibitors
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5H08
 
 | | Human PTPRZ D1 domain complexed with NAZ2329 | | Descriptor: | 3-{[2-Ethoxy-5-(trifluoromethyl)benzyl]sulfanyl}-N-(phenylsulfonyl)thiophene-2-carboxamide, Receptor-type tyrosine-protein phosphatase zeta | | Authors: | Sugawara, H. | | Deposit date: | 2016-10-04 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Targeting PTPRZ inhibits stem cell-like properties and tumorigenicity in glioblastoma cells
Sci Rep, 7, 2017
|
|
6AHS
 
 | | Mouse Kallikrein 7 in complex with imidazolinylindole derivative | | Descriptor: | 1-[(2-chlorophenyl)sulfonyl]-5-methyl-3-[(4R)-2-methyl-4,5-dihydro-1H-imidazol-4-yl]-1H-indole, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Sugawara, H. | | Deposit date: | 2018-08-20 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery and structure-activity relationship of imidazolinylindole derivatives as kallikrein 7 inhibitors.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
5YJP
 
 | | Human chymase in complex with 3-(ethoxyimino)-7-oxo-1,4-diazepane derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-((R)-1-((R,Z)-6-(5-chloro-2-methoxybenzyl)-3-(ethoxyimino)-7-oxo-1,4-diazepane-1-carboxamido)propyl)benzoic acid, ZINC ION, ... | | Authors: | Sugawara, H. | | Deposit date: | 2017-10-11 | | Release date: | 2017-12-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design, synthesis, and binding mode analysis of novel and potent chymase inhibitors
Bioorg. Med. Chem. Lett., 28, 2018
|
|
5ZFI
 
 | | Mouse kallikrein 7 in complex with 6-benzyl-1,4-diazepan-7-one derivative | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[(3Z,6R)-6-[(2,6-dichlorophenyl)methyl]-3-(dimethylhydrazinylidene)-7-oxo-1,4-diazepan-1-yl]-N-[3-(1-methyl-1H-pyrazol-4-yl)phenyl]acetamide, Kallikrein-7 | | Authors: | Sugawara, H. | | Deposit date: | 2018-03-06 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based drug design to overcome species differences in kallikrein 7 inhibition of 1,3,6-trisubstituted 1,4-diazepan-7-ones.
Bioorg. Med. Chem., 26, 2018
|
|
5ZFH
 
 | | Mouse Kallikrein 7 | | Descriptor: | Kallikrein-7 | | Authors: | Sugawara, H. | | Deposit date: | 2018-03-06 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure-based drug design to overcome species differences in kallikrein 7 inhibition of 1,3,6-trisubstituted 1,4-diazepan-7-ones.
Bioorg. Med. Chem., 26, 2018
|
|
5YJK
 
 | | Human kallikrein 7 in complex with 1,4-diazepane-7-one 1-acetamide derivative | | Descriptor: | (R)-2-(6-(5-chloro-2-methoxybenzyl)-3-(2,2-dimethylhydrazono)-7-oxo-1,4-diazepan-1-yl)-N-(3-(methylsulfonyl)phenyl)acetamide, CHLORIDE ION, Kallikrein-7 | | Authors: | Sugawara, H. | | Deposit date: | 2017-10-11 | | Release date: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery and structure-activity relationship study of 1,3,6-trisubstituted 1,4-diazepane-7-ones as novel human kallikrein 7 inhibitors
Bioorg. Med. Chem. Lett., 27, 2017
|
|
1BWV
 
 | | Activated Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase (RUBISCO) Complexed with the Reaction Intermediate Analogue 2-Carboxyarabinitol 1,5-Bisphosphate | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE) | | Authors: | Sugawara, H, Yamamoto, H, Shibata, N, Inoue, T, Miyake, C, Yokota, A, Kai, Y. | | Deposit date: | 1998-09-29 | | Release date: | 1999-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of carboxylase reaction-oriented ribulose 1, 5-bisphosphate carboxylase/oxygenase from a thermophilic red alga, Galdieria partita.
J.Biol.Chem., 274, 1999
|
|
1WMZ
 
 | | Crystal Structure of C-type Lectin CEL-I complexed with N-acetyl-D-galactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Sugawara, H, Kusunoki, M, Kurisu, G, Fujimoto, T, Aoyagi, H, Hatakeyama, T. | | Deposit date: | 2004-07-22 | | Release date: | 2004-09-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characteristic Recognition of N-Acetylgalactosamine by an Invertebrate C-type Lectin, CEL-I, Revealed by X-ray Crystallographic Analysis
J.Biol.Chem., 279, 2004
|
|
1WMY
 
 | | Crystal Structure of C-type Lectin CEL-I from Cucumaria echinata | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, lectin CEL-I, ... | | Authors: | Sugawara, H, Kusunoki, M, Kurisu, G, Fujimoto, T, Aoyagi, H, Hatakeyama, T. | | Deposit date: | 2004-07-22 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characteristic Recognition of N-Acetylgalactosamine by an Invertebrate C-type Lectin, CEL-I, Revealed by X-ray Crystallographic Analysis
J.Biol.Chem., 279, 2004
|
|
1WN0
 
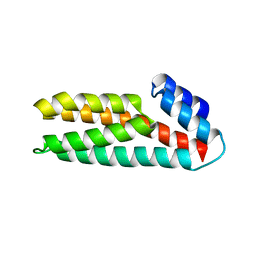 | | Crystal Structure of Histidine-containing Phosphotransfer Protein, ZmHP2, from maize | | Descriptor: | histidine-containing phosphotransfer protein | | Authors: | Sugawara, H, Kawano, Y, Hatakeyama, T, Yamaya, T, Kamiya, N, Sakakibara, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-24 | | Release date: | 2005-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the histidine-containing phosphotransfer protein ZmHP2 from maize
Protein Sci., 14, 2005
|
|
5YJM
 
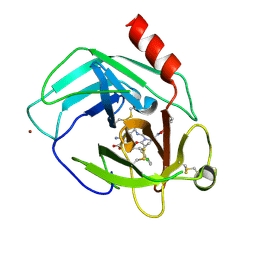 | | Human chymase in complex with 7-oxo-3-(phenoxyimino)-1,4-diazepane derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-4-((R)-1-((R,Z)-6-(5-chloro-2-methoxybenzyl)-7-oxo-3-(phenoxyimino)-1,4-diazepane-1-carboxamido)propyl)benzoic acid, ZINC ION, ... | | Authors: | Sugawara, H. | | Deposit date: | 2017-10-11 | | Release date: | 2017-12-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design, synthesis, and binding mode analysis of novel and potent chymase inhibitors
Bioorg. Med. Chem. Lett., 28, 2018
|
|
1BYO
 
 | | WILD-TYPE PLASTOCYANIN FROM SILENE | | Descriptor: | COPPER (II) ION, PROTEIN (PLASTOCYANIN) | | Authors: | Sugawara, H, Inoue, T, Li, C, Gotowda, M, Hibino, T, Takabe, T, Kai, Y. | | Deposit date: | 1998-10-19 | | Release date: | 1999-10-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of wild-type and mutant plastocyanins from a higher plant, Silene.
J.Biochem.(Tokyo), 125, 1999
|
|
1BYP
 
 | | E43K,D44K DOUBLE MUTANT PLASTOCYANIN FROM SILENE | | Descriptor: | COPPER (II) ION, PROTEIN (PLASTOCYANIN) | | Authors: | Sugawara, H, Inoue, T, Li, C, Gotowda, M, Hibino, T, Takabe, T, Kai, Y. | | Deposit date: | 1998-10-19 | | Release date: | 1999-10-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of wild-type and mutant plastocyanins from a higher plant, Silene.
J.Biochem.(Tokyo), 125, 1999
|
|
5AWX
 
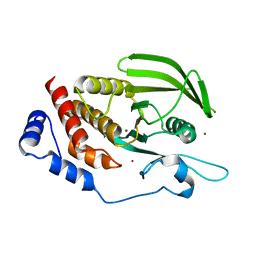 | | Crystal structure of Human PTPRZ D1 domain | | Descriptor: | BROMIDE ION, Receptor-type tyrosine-protein phosphatase zeta | | Authors: | Sugawara, H. | | Deposit date: | 2015-07-10 | | Release date: | 2016-02-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Small-molecule inhibition of PTPRZ reduces tumor growth in a rat model of glioblastoma
Sci Rep, 6, 2016
|
|
5KGN
 
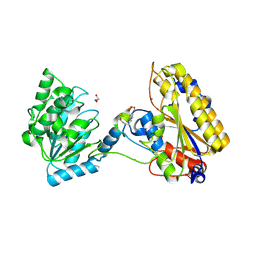 | | 1.95A resolution structure of independent phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (2d) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
5KGM
 
 | | 2.95A resolution structure of Apo independent phosphoglycerate mutase from C. elegans (monoclinic form) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
5KGL
 
 | | 2.45A resolution structure of Apo independent phosphoglycerate mutase from C. elegans (orthorhombic form) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
7QPB
 
 | | Catalytic C-lobe of the HECT-type ubiquitin ligase E6AP in complex with a hybrid foldamer-peptide macrocycle | | Descriptor: | Isoform I of Ubiquitin-protein ligase E3A, hybrid foldamer-peptide macrocycle | | Authors: | Dengler, S, Howard, R.T, Morozov, V, Tsiamantas, C, Douat, C, Suga, H, Huc, I. | | Deposit date: | 2022-01-03 | | Release date: | 2023-09-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | Display Selection of a Hybrid Foldamer-Peptide Macrocycle.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6WGN
 
 | | Crystal structure of K-Ras(G12D) GppNHp bound to cyclic peptide ligand KD2 | | Descriptor: | Cyclic Peptide KD2, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Zhang, Z, Gao, R, Hu, Q, Peacock, H, Peacock, D.M, Shokat, K.M, Suga, H. | | Deposit date: | 2020-04-06 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | GTP-State-Selective Cyclic Peptide Ligands of K-Ras(G12D) Block Its Interaction with Raf.
Acs Cent.Sci., 6, 2020
|
|
8JE4
 
 | | Crystal structure of LimF prenyltransferase (H239G/W273T mutant) bound with the thiodiphosphate moiety of farnesyl S-thiolodiphosphate (FSPP) | | Descriptor: | MAGNESIUM ION, TRIHYDROGEN THIODIPHOSPHATE, prenyltransferase, ... | | Authors: | Hamada, K, Oguni, A, Zhang, Y, Satake, M, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Switching Prenyl Donor Specificities of Cyanobactin Prenyltransferases.
J.Am.Chem.Soc., 145, 2023
|
|
8WM0
 
 | | Crystal structure of TNIK-thiopeptide wTP3 complex | | Descriptor: | ADENOSINE, THIOPEPTIDE wTP3, TRAF2 and NCK-interacting protein kinase | | Authors: | Hamada, K, Kobayashi, S, Vinogradov, A.A, Zhang, Y, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2023-10-01 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Compact Reprogrammed Genetic Code for De Novo Discovery of Proteolytically Stable Thiopeptides.
J.Am.Chem.Soc., 2024
|
|
8YE0
 
 | | Crystal structure of KgpF prenyltransferase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LynF/TruF/PatF family peptide O-prenyltransferase, MAGNESIUM ION, ... | | Authors: | Hamada, K, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2024-02-21 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | De Novo Discovery of Pseudo-Natural Prenylated Macrocyclic Peptide Ligands.
Angew.Chem.Int.Ed.Engl., 2024
|
|
7TL8
 
 | | 1.95A resolution structure of independent phosphoglycerate mutase from S. aureus in complex with a macrocyclic peptide inhibitor (Sa-D3) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, MANGANESE (II) ION, Peptide Sa-D3 | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Dranchak, P, Queme, B, Aitha, M, van Neer, R.H.P, Kimura, H, Katho, T, Suga, H, Inglese, J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Serum-Stable and Selective Backbone-N-Methylated Cyclic Peptides That Inhibit Prokaryotic Glycolytic Mutases.
Acs Chem.Biol., 17, 2022
|
|
