6B2R
 
 | | Crystal structure of Xanthomonas campestris OleA H285A | | Descriptor: | 3-oxoacyl-[ACP] synthase III, GLYCEROL | | Authors: | Jensen, M.R, Goblirsch, B.R, Esler, M.A, Christenson, J.K, Mohamed, F.A, Wackett, L.P, Wilmot, C.M. | | Deposit date: | 2017-09-20 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The role of OleA His285 in orchestration of long-chain acyl-coenzyme A substrates.
FEBS Lett., 592, 2018
|
|
6B2T
 
 | | Crystal structure of Xanthomonas campestris OleA H285D | | Descriptor: | 3-oxoacyl-[ACP] synthase III, GLYCEROL, PHOSPHATE ION | | Authors: | Jensen, M.R, Goblirsch, B.R, Esler, M.A, Christenson, J.K, Mohamed, F.A, Wackett, L.P, Wilmot, C.M. | | Deposit date: | 2017-09-20 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The role of OleA His285 in orchestration of long-chain acyl-coenzyme A substrates.
FEBS Lett., 592, 2018
|
|
6B3X
 
 | | Crystal structure of CstF-50 in complex with CstF-77 | | Descriptor: | Cleavage stimulation factor subunit 1, Cleavage stimulation factor subunit 3 | | Authors: | Yang, W, Hsu, P, Yang, F, Song, J.E, Varani, G. | | Deposit date: | 2017-09-25 | | Release date: | 2017-11-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reconstitution of the CstF complex unveils a regulatory role for CstF-50 in recognition of 3'-end processing signals.
Nucleic Acids Res., 46, 2018
|
|
1IKI
 
 | | COMPLEX OF STREPTOMYCES R61 DD-PEPTIDASE WITH THE PRODUCTS OF A SPECIFIC PEPTIDOGLYCAN SUBSTRATE FRAGMENT | | Descriptor: | D-ALANINE, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, GLYCYL-L-ALPHA-AMINO-EPSILON-PIMELYL-D-ALANINE | | Authors: | Mcdonough, M.A, Anderson, J.W, Silvaggi, N.R, Pratt, R.F, Knox, J.R, Kelly, J.A. | | Deposit date: | 2001-05-03 | | Release date: | 2002-09-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structures of two kinetic intermediates reveal species specificity of penicillin-binding proteins.
J.Mol.Biol., 322, 2002
|
|
1IKG
 
 | | MICHAELIS COMPLEX OF STREPTOMYCES R61 DD-PEPTIDASE WITH A SPECIFIC PEPTIDOGLYCAN SUBSTRATE FRAGMENT | | Descriptor: | D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, GLYCYL-L-ALPHA-AMINO-EPSILON-PIMELYL-D-ALANYL-D-ALANINE | | Authors: | Mcdonough, M.A, Anderson, J.W, Silvaggi, N.R, Pratt, R.F, Knox, J.R, Kelly, J.A. | | Deposit date: | 2001-05-03 | | Release date: | 2002-09-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of two kinetic intermediates reveal species specificity of penicillin-binding proteins.
J.Mol.Biol., 322, 2002
|
|
4CAV
 
 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a benzofuran ligand R0-09-4879 | | Descriptor: | 3-[[3-methyl-2-[[2,3,4-tris(fluoranyl)phenoxy]methyl]-1-benzofuran-4-yl]oxy]-N-(pyridin-3-ylmethyl)propan-1-amine, CHLORIDE ION, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Robinson, D.A, Fang, W, Raimi, O.G, Blair, D.E, Harrison, J, Ruda, G.F, Lockhart, D.E.A, Torrie, L.S, Wyatt, P.G, Gilbert, I.H, Van Aalten, D.M.F. | | Deposit date: | 2013-10-09 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | N-Myristoyltransferase is a Cell Wall Target in Aspergillus Fumigatus.
Acs Chem.Biol., 10, 2015
|
|
8RNU
 
 | |
4CAX
 
 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a pyrazole sulphonamide ligand (DDD85646) | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Raimi, O.G, Robinson, D.A, Fang, W, Blair, D.E, Harrison, J, Ruda, G.F, Lockhart, D.E.A, Torrie, L.S, Wyatt, P.G, Gilbert, I.H, Van Aalten, D.M.F. | | Deposit date: | 2013-10-09 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N-Myristoyltransferase is a Cell Wall Target in Aspergillus Fumigatus.
Acs Chem.Biol., 10, 2015
|
|
8W2W
 
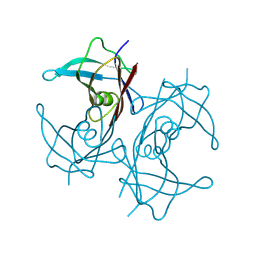 | | Structure of transthyretin synthetic mutation A120L | | Descriptor: | Transthyretin | | Authors: | Yang, K, Sun, X, Ferguson, J.A, Stanfield, R.L, Wright, P.E. | | Deposit date: | 2024-02-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Mispacking of the F87 sidechain drives aggregation-promoting conformational fluctuations in the subunit interfaces of the transthyretin tetramer.
Protein Sci., 33, 2024
|
|
6CWZ
 
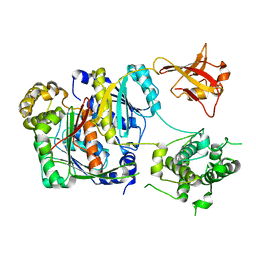 | | Crystal structure of apo SUMO E1 | | Descriptor: | SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ZINC ION | | Authors: | Lv, Z, Yuan, L, Atkison, J.H, Williams, K.M, Olsen, S.K. | | Deposit date: | 2018-04-01 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular mechanism of a covalent allosteric inhibitor of SUMO E1 activating enzyme.
Nat Commun, 9, 2018
|
|
8T1U
 
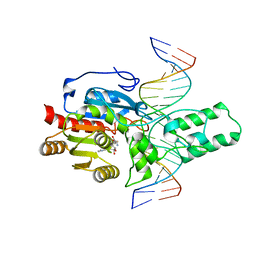 | | Crystal structure of the DRM2-CTA DNA complex | | Descriptor: | DNA (5'-D(P*AP*TP*TP*AP*TP*TP*AP*AP*TP*(C49)P*TP*AP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(P*TP*AP*AP*AP*TP*TP*TP*AP*GP*AP*TP*TP*AP*AP*TP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | Authors: | Chen, J, Lu, J, Song, J. | | Deposit date: | 2023-06-03 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | DNA conformational dynamics in the context-dependent non-CG CHH methylation by plant methyltransferase DRM2.
J.Biol.Chem., 299, 2023
|
|
8T5G
 
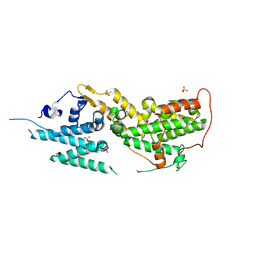 | | SOS2 co-crystal structure with fragment bound (compound 12) | | Descriptor: | DIMETHYL SULFOXIDE, SULFATE ION, Son of sevenless homolog 2, ... | | Authors: | Gunn, R.J, Lawson, J.D, Ivetac, A, Ulaganathan, T, Coulombe, R, Fethiere, J. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
8T5M
 
 | | SOS2 crystal structure with fragment bound (compound 14) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(1R,2S)-1-hydroxy-2-{[2-(4-hydroxyphenyl)ethyl]amino}propyl]phenol, SULFATE ION, ... | | Authors: | Gunn, R.J, Lawson, J.D, Ivetac, A, Ulaganathan, T, Coulombe, R, Fethiere, J. | | Deposit date: | 2023-06-14 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
8T5R
 
 | | SOS2 crystal structure with fragment bound (compound 13) | | Descriptor: | 4-(aminomethyl)benzene-1-sulfonamide, SULFATE ION, Son of sevenless homolog 2 | | Authors: | Gunn, R.J, Lawson, J.D, Ivetac, A, Ulaganathan, T, Coulombe, R, Fethiere, J. | | Deposit date: | 2023-06-14 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Discovery of Five SOS2 Fragment Hits with Binding Modes Determined by SOS2 X-Ray Cocrystallography.
J.Med.Chem., 67, 2024
|
|
8T12
 
 | |
8T13
 
 | |
1W0O
 
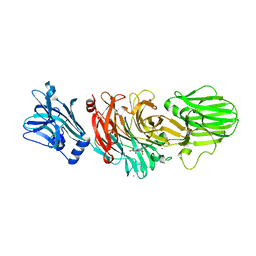 | | Vibrio cholerae sialidase | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Moustafa, I, Connaris, H, Taylor, M, Zaitsev, V, Wilson, J.C, Kiefel, M.J, von-Itzstein, M, Taylor, G. | | Deposit date: | 2004-06-09 | | Release date: | 2004-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sialic Acid Recognition by Vibrio Cholerae Neuraminidase
J.Biol.Chem., 279, 2004
|
|
4NQA
 
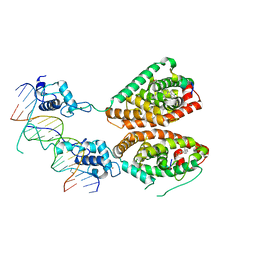 | | Crystal structure of liganded hRXR-alpha/hLXR-beta heterodimer on DNA | | Descriptor: | (9cis)-retinoic acid, 5'-D(*TP*AP*AP*GP*GP*TP*CP*AP*CP*TP*TP*CP*AP*GP*GP*TP*CP*A)-3', 5'-D(*TP*AP*TP*GP*AP*CP*CP*TP*GP*AP*AP*GP*TP*GP*AP*CP*CP*T)-3', ... | | Authors: | Lou, X.H, Toresson, G, Benod, C, Suh, J.H, Phillips, K.J, Webb, P, Gustafsson, J.A. | | Deposit date: | 2013-11-24 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structure of the retinoid X receptor alpha-liver X receptor beta (RXR alpha-LXR beta ) heterodimer on DNA.
Nat.Struct.Mol.Biol., 21, 2014
|
|
6CWY
 
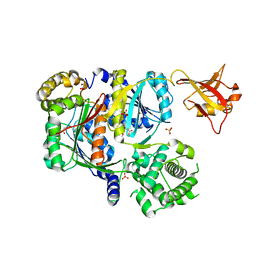 | | Crystal structure of SUMO E1 in complex with an allosteric inhibitor | | Descriptor: | GLYCEROL, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Lv, Z, Yuan, L, Atkison, J.H, Williams, K.M, Olsen, S.K. | | Deposit date: | 2018-04-01 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.462 Å) | | Cite: | Molecular mechanism of a covalent allosteric inhibitor of SUMO E1 activating enzyme.
Nat Commun, 9, 2018
|
|
4PRF
 
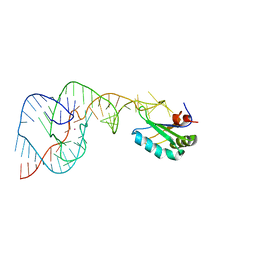 | | A Second Look at the HDV Ribozyme Structure and Dynamics. | | Descriptor: | Hepatitis Delta virus ribozyme, STRONTIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Kapral, G.J, Jain, S, Noeske, J, Doudna, J.A, Richardson, D.C, Richardson, J.S. | | Deposit date: | 2014-03-05 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | New tools provide a second look at HDV ribozyme structure, dynamics and cleavage.
Nucleic Acids Res., 42, 2014
|
|
5E7C
 
 | | Macromolecular diffractive imaging using imperfect crystals - Bragg data | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ayyer, K, Yefanov, O, Oberthuer, D, Roy-Chowdhury, S, Galli, L, Mariani, V, Basu, S, Coe, J, Conrad, C.E, Fromme, R, Schaffner, A, Doerner, K, James, D, Kupitz, C, Metz, M, Nelson, G, Xavier, P.L, Beyerlein, K.R, Schmidt, M, Sarrou, I, Spence, J.C.H, Weierstall, U, White, T.A, Yang, J.-H, Zhao, Y, Liang, M, Aquila, A, Hunter, M.S, Robinson, J.S, Koglin, J.E, Boutet, S, Fromme, P, Barty, A, Chapman, H.N. | | Deposit date: | 2015-10-12 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Macromolecular diffractive imaging using imperfect crystals.
Nature, 530, 2016
|
|
1S1K
 
 | |
8TAR
 
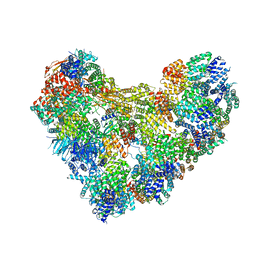 | | APC/C-CDH1-UBE2C-Ubiquitin-CyclinB-NTD | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Bodrug, T, Welsh, K.A, Bolhuis, D.L, Paulakonis, E, Martinez-Chacin, R.C, Liu, B, Pinkin, N, Bonacci, T, Cui, L, Xu, P, Roscow, O, Amann, S.J, Grishkovskaya, I, Emanuele, M.J, Harrison, J.S, Steimel, J.P, Hahn, K.M, Zhang, W, Zhong, E, Haselbach, D, Brown, N.G. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Time-resolved cryo-EM (TR-EM) analysis of substrate polyubiquitination by the RING E3 anaphase-promoting complex/cyclosome (APC/C).
Nat.Struct.Mol.Biol., 30, 2023
|
|
8U4U
 
 | |
8TAU
 
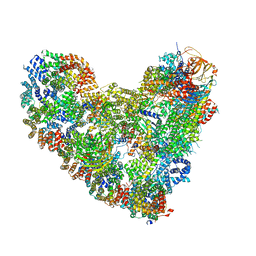 | | APC/C-CDH1-UBE2C-UBE2S-Ubiquitin-CyclinB | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Bodrug, T, Welsh, K.A, Bolhuis, D.L, Paulakonis, E, Martinez-Chacin, R.C, Liu, B, Pinkin, N, Bonacci, T, Cui, L, Xu, P, Roscow, O, Amann, S.J, Grishkovskaya, I, Emanuele, M.J, Harrison, J.S, Steimel, J.P, Hahn, K.M, Zhang, W, Zhong, E, Haselbach, D, Brown, N.G. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Time-resolved cryo-EM (TR-EM) analysis of substrate polyubiquitination by the RING E3 anaphase-promoting complex/cyclosome (APC/C).
Nat.Struct.Mol.Biol., 30, 2023
|
|
