7KPG
 
 | | Blocking Fab 25 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | Descriptor: | Heavy chain of Fab 25 anti-SIRP-alpha antibody, Light chain of Fab 25 anti-SIRP-alpha antibody, SULFATE ION, ... | | Authors: | Sim, J, Pons, J. | | Deposit date: | 2020-11-11 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Targeting the myeloid checkpoint receptor SIRP alpha potentiates innate and adaptive immune responses to promote anti-tumor activity.
J Hematol Oncol, 13, 2020
|
|
4DAW
 
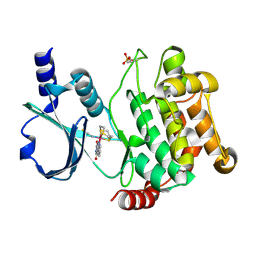 | | Crystal structure of PAK1 kinase domain with the ruthenium phthalimide complex | | Descriptor: | Serine/threonine-protein kinase PAK 1, [1,3-dioxo-6-(pyridin-2-yl-kappaN)-2,3-dihydro-1H-isoindol-5-yl-kappaC~5~][(thioxomethylidene)azanido-kappaN](1,4,7-trithionane-kappa~3~S~1~,S~4~,S~7~)ruthenium | | Authors: | Maksimoska, J, Marmorstein, R. | | Deposit date: | 2012-01-13 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The art of filling protein pockets efficiently with octahedral metal complexes.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
8UGT
 
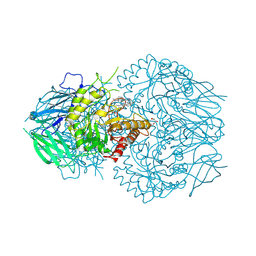 | | E. eligens beta-glucuronidase bound to UNC10206581-G | | Descriptor: | 8-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-5-(methylamino)-1,2,3,4-tetrahydro[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinoline, Beta-glucuronidase, GLYCEROL, ... | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2023-10-06 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advanced piperazine-containing inhibitors target microbial beta-glucuronidases linked to gut toxicity.
Rsc Chem Biol, 5, 2024
|
|
8GEN
 
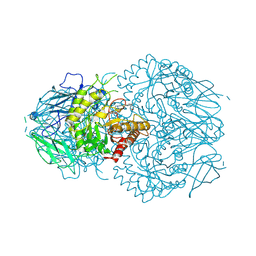 | | E. eligens beta-glucuronidase bound to UNC10201652-glucuronide | | Descriptor: | 8-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-5-(morpholin-4-yl)-1,2,3,4-tetrahydro[1,2,3]triazino[4',5':4,5]thieno[2,3 -c]isoquinoline, Beta-glucuronidase | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
8GEQ
 
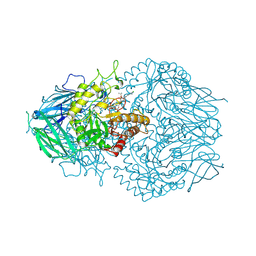 | | E. eligens beta-glucuronidase bound to ceritinib-glucuronide | | Descriptor: | 4-amino-5-chloro-2-{4-(1-beta-D-glucopyranuronosylpiperidin-4-yl)-5-methyl-2-[(propan-2-yl)oxy]anilino}pyrimidine, Beta-glucuronidase | | Authors: | Simpson, J.B, Kowalewski, M.K, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
8GES
 
 | | R. hominis 2 beta-glucuronidase bound to UNC10201652-glucuronide | | Descriptor: | 8-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-5-(morpholin-4-yl)-1,2,3,4-tetrahydro[1,2,3]triazino[4',5':4,5]thieno[2,3 -c]isoquinoline, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-02-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
8GER
 
 | | E. eligens beta-glucuronidase bound to norquetiapine-glucuronide | | Descriptor: | 11-(4-beta-D-glucopyranuronosylpiperazin-1-yl)dibenzo[b,f][1,4]thiazepine, Beta-glucuronidase | | Authors: | Simpson, J.B, Lietzan, A.D, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
8GEO
 
 | | E. eligens beta-glucuronidase bound to 3-OH-desloratidine-glucuronide | | Descriptor: | 8-chloro-11-(1-beta-D-glucopyranuronosylpiperidin-4-ylidene)-3-hydroxy-6,11-dihydro-5H-benzo[5,6]cyclohepta[1,2-b]pyridine, Beta-glucuronidase, GLYCEROL | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
8GET
 
 | | R. hominis 2 beta-glucuronidase bound to norquetiapine-glucuronide | | Descriptor: | 11-(4-beta-D-glucopyranuronosylpiperazin-1-yl)dibenzo[b,f][1,4]thiazepine, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
7KGY
 
 | | Beta-glucuronidase from Faecalibacterium prausnitzii bound to the inhibitor UNC10201652-glucuronide | | Descriptor: | 8-(4-beta-D-glucopyranuronosylpiperazin-1-yl)-5-(morpholin-4-yl)-1,2,3,4-tetrahydro[1,2,3]triazino[4',5':4,5]thieno[2,3 -c]isoquinoline, Beta-glucuronidase, GLYCEROL | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2020-10-19 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Microbial enzymes induce colitis by reactivating triclosan in the mouse gastrointestinal tract.
Nat Commun, 13, 2022
|
|
6G5R
 
 | |
6G5S
 
 | | Solution structure of the TPR domain of the cell division coordinator, CpoB | | Descriptor: | Cell division coordinator CpoB | | Authors: | Simorre, J.P, Maya Martinez, R.C, Bougault, C, Vollmer, W, Egan, A. | | Deposit date: | 2018-03-29 | | Release date: | 2018-08-08 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Induced conformational changes activate the peptidoglycan synthase PBP1B.
Mol. Microbiol., 110, 2018
|
|
6FZK
 
 | | NMR structure of UB2H, regulatory domain of PBP1b from E. coli | | Descriptor: | Penicillin-binding protein 1B | | Authors: | Simorre, J.P, Maya Martinez, R.C, Bougault, C, Egan, A.J.F, Vollmer, W. | | Deposit date: | 2018-03-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Induced conformational changes activate the peptidoglycan synthase PBP1B.
Mol. Microbiol., 110, 2018
|
|
5DFP
 
 | | Crystal structure of PAK1 in complex with an inhibitor compound FRAX1036 | | Descriptor: | 6-[2-chloro-4-(6-methylpyrazin-2-yl)phenyl]-8-ethyl-2-{[2-(1-methylpiperidin-4-yl)ethyl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, DIMETHYL SULFOXIDE, Serine/threonine-protein kinase PAK 1 | | Authors: | Maksimoska, J, Marmorstein, R, Wang, W. | | Deposit date: | 2015-08-27 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of Selective PAK1 Inhibitor G-5555: Improving Properties by Employing an Unorthodox Low-pK a Polar Moiety.
Acs Med.Chem.Lett., 6, 2015
|
|
4EQC
 
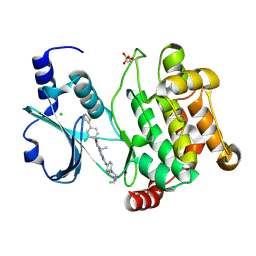 | | Crystal structure of PAK1 kinase domain in complex with FRAX597 inhibitor | | Descriptor: | 6-[2-chloro-4-(1,3-thiazol-5-yl)phenyl]-8-ethyl-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, CHLORIDE ION, Serine/threonine-protein kinase PAK 1 | | Authors: | Maksimoska, J, Marmorstein, R. | | Deposit date: | 2012-04-18 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | FRAX597, a Small Molecule Inhibitor of the p21-activated Kinases, Inhibits Tumorigenesis of Neurofibromatosis Type 2 (NF2)-associated Schwannomas.
J.Biol.Chem., 288, 2013
|
|
4PZT
 
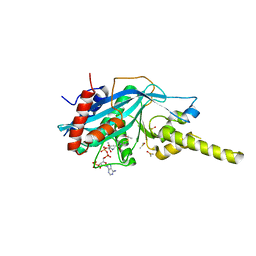 | | Crystal structure of p300 histone acetyltransferase domain in complex with an inhibitor, Acetonyl-Coenzyme A | | Descriptor: | DIMETHYL SULFOXIDE, Histone acetyltransferase p300, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-3-HYDROXY-2,2-DIMETHYL-4-OXO-4-{[3-OXO-3-({2-[(2-OXOPROPYL)THIO]ETHYL}AMINO)PROPYL]AMINO}BUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Maksimoska, J, Marmorstein, R. | | Deposit date: | 2014-03-31 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the p300 Histone Acetyltransferase Bound to Acetyl-Coenzyme A and Its Analogues.
Biochemistry, 53, 2014
|
|
3BWF
 
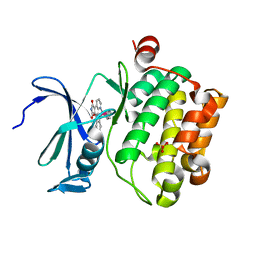 | | Crystal structure of the human Pim1 in complex with an osmium compound | | Descriptor: | PYRIDOCARBAZOLE CYCLOPENTADIENYL OS(CO) COMPLEX, Proto-oncogene serine/threonine-protein kinase Pim-1, SULFATE ION | | Authors: | Maksimoska, J, Filippakopoulos, P, Knapp, S, Meggers, E. | | Deposit date: | 2008-01-09 | | Release date: | 2008-06-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Similar biological activities of two isostructural ruthenium and osmium complexes.
Chemistry, 14, 2008
|
|
4PZR
 
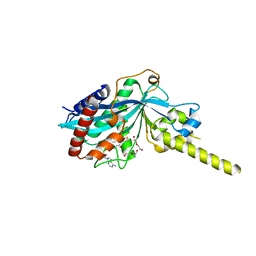 | |
4PZS
 
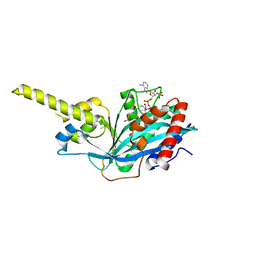 | |
3FY0
 
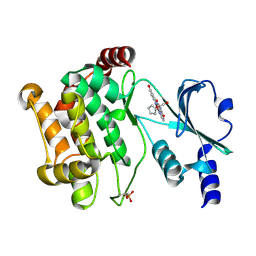 | |
3FXZ
 
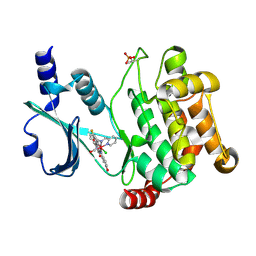 | |
1Z2M
 
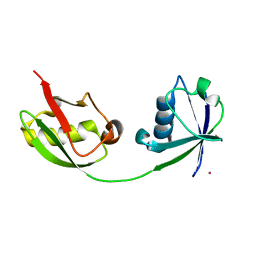 | | Crystal Structure of ISG15, the Interferon-Induced Ubiquitin Cross Reactive Protein | | Descriptor: | OSMIUM 4+ ION, interferon, alpha-inducible protein (clone IFI-15K) | | Authors: | Narasimhan, J, Wang, M, Fu, Z, Klein, J.M, Haas, A.L, Kim, J.J. | | Deposit date: | 2005-03-08 | | Release date: | 2005-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Interferon-induced Ubiquitin-like Protein ISG15.
J.Biol.Chem., 280, 2005
|
|
2OI4
 
 | |
2L56
 
 | |
6NMV
 
 | | Non-Blocking Fab 218 anti-SIRP-alpha antibody in complex with SIRP-alpha Variant 1 | | Descriptor: | Fab 218 anti-SIRP-alpha antibody Variable Heavy Chain, Fab 218 anti-SIRP-alpha antibody Variable Light Chain, Tyrosine-protein phosphatase non-receptor type substrate 1 | | Authors: | Wibowo, A.S, Carter, J.J, Sim, J. | | Deposit date: | 2019-01-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of high affinity, pan-allelic, and pan-mammalian reactive antibodies against the myeloid checkpoint receptor SIRP alpha.
Mabs, 11, 2019
|
|
