4YIO
 
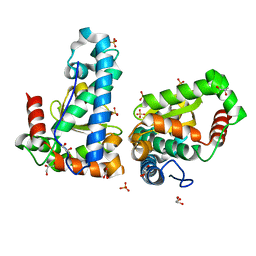 | | X-ray structure of the iron/manganese cambialistic superoxide dismutase from Streptococcus thermophilus | | Descriptor: | FE (III) ION, GLYCEROL, SULFATE ION, ... | | Authors: | Russo Krauss, I, Merlino, A, Pica, A, Sica, F. | | Deposit date: | 2015-03-02 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fine tuning of metal-specific activity in the Mn-like group of cambialistic superoxide dismutases
Rsc Adv, 5, 2015
|
|
4YIP
 
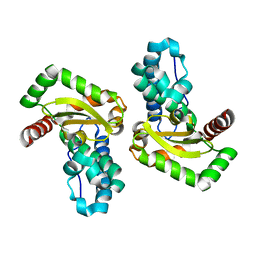 | | X-ray structure of the iron/manganese cambialistic superoxide dismutase from Streptococcus mutans | | Descriptor: | FE (III) ION, Superoxide dismutase [Mn/Fe] | | Authors: | Russo Krauss, I, Merlino, A, Pica, A, Sica, F. | | Deposit date: | 2015-03-02 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fine tuning of metal-specific activity in the Mn-like group of cambialistic superoxide dismutases
Rsc Adv, 5, 2015
|
|
4DII
 
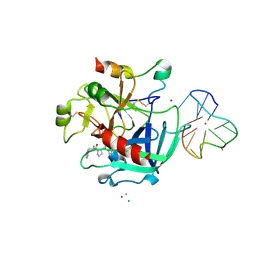 | | X-ray structure of the complex between human alpha thrombin and thrombin binding aptamer in the presence of potassium ions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2012-01-31 | | Release date: | 2012-07-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity.
Nucleic Acids Res., 40, 2012
|
|
4DIH
 
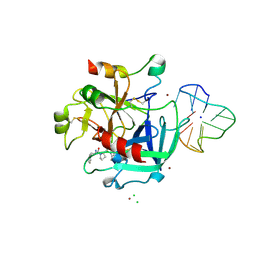 | | X-ray structure of the complex between human alpha thrombin and thrombin binding aptamer in the presence of sodium ions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2012-01-31 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity.
Nucleic Acids Res., 40, 2012
|
|
7NTU
 
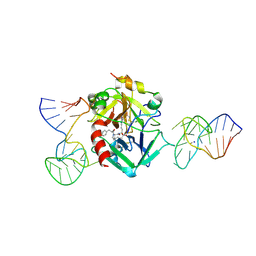 | | X-ray structure of the complex between human alpha thrombin and two duplex/quadruplex aptamers: NU172 and HD22_27mer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, HD22_27mer, ... | | Authors: | Troisi, R, Santamaria, A, Sica, F. | | Deposit date: | 2021-03-10 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and functional analysis of the simultaneous binding of two duplex/quadruplex aptamers to human alpha-thrombin.
Int.J.Biol.Macromol., 181, 2021
|
|
3F8G
 
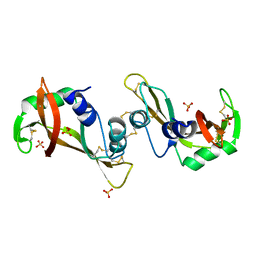 | | The X-ray structure of a dimeric variant of human pancreatic ribonuclease with high cytotoxic and antitumor activities | | Descriptor: | Ribonuclease pancreatic, SULFATE ION | | Authors: | Merlino, A, Avella, G, Mazzarella, L, Sica, F. | | Deposit date: | 2008-11-12 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural features for the mechanism of antitumor action of a dimeric human pancreatic ribonuclease variant.
Protein Sci., 18, 2009
|
|
3FKZ
 
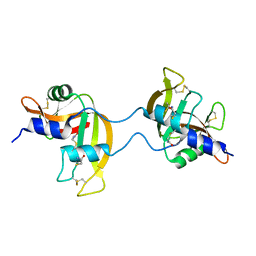 | | X-ray structure of the non covalent swapped form of the S16G/T17N/A19P/A20S/K31C/S32C mutant of bovine pancreatic ribonuclease | | Descriptor: | Ribonuclease pancreatic | | Authors: | Merlino, A, Russo Krauss, I, Perillo, M, Mattia, C.A, Ercole, C, Picone, D, Vergara, A, Sica, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Toward an antitumor form of bovine pancreatic ribonuclease: The crystal structure of three noncovalent dimeric mutants
Biopolymers, 91, 2009
|
|
3FL3
 
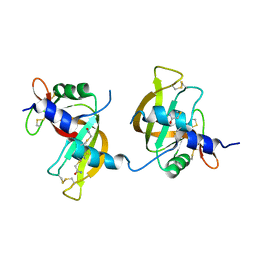 | | X-ray structure of the ligand free non covalent swapped form of the A19P/Q28L/K31C/S32C mutant of bovine pancreatic ribonuclease | | Descriptor: | Ribonuclease pancreatic, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Merlino, A, Russo Krauss, I, Perillo, M, Mattia, C.A, Ercole, C, Picone, D, Vergara, A, Sica, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Toward an antitumor form of bovine pancreatic ribonuclease: The crystal structure of three noncovalent dimeric mutants
Biopolymers, 91, 2009
|
|
3FL0
 
 | | X-ray structure of the non covalent swapped form of the Q28L/K31C/S32C mutant of bovine pancreatic ribonuclease in complex with 2'-DEOXYCYTIDINE-2'-DEOXYGUANOSINE-3',5'-MONOPHOSPHATE | | Descriptor: | 2'-DEOXYCYTIDINE-2'-DEOXYGUANOSINE-3',5'-MONOPHOSPHATE, Ribonuclease pancreatic | | Authors: | Merlino, A, Russo Krauss, I, Perillo, M, Mattia, C.A, Ercole, C, Picone, D, Vergara, A, Sica, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Toward an antitumor form of bovine pancreatic ribonuclease: The crystal structure of three noncovalent dimeric mutants
Biopolymers, 91, 2009
|
|
3FL1
 
 | | X-ray structure of the non covalent swapped form of the A19P/Q28L/K31C/S32C mutant of bovine pancreatic ribonuclease in complex with 2'-DEOXYCYTIDINE-2'-DEOXYGUANOSINE-3',5'-MONOPHOSPHATE | | Descriptor: | 2'-DEOXYCYTIDINE-2'-DEOXYGUANOSINE-3',5'-MONOPHOSPHATE, Ribonuclease pancreatic, SULFATE ION, ... | | Authors: | Merlino, A, Russo Krauss, I, Perillo, M, Mattia, C.A, Ercole, C, Picone, D, Vergara, A, Sica, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Toward an antitumor form of bovine pancreatic ribonuclease: The crystal structure of three noncovalent dimeric mutants
Biopolymers, 91, 2009
|
|
1NVG
 
 | | N249Y MUTANT OF THE ALCOHOL DEHYDROGENASE FROM THE ARCHAEON SULFOLOBUS SOLFATARICUS-TETRAGONAL CRYSTAL FORM | | Descriptor: | NAD-dependent alcohol dehydrogenase, ZINC ION | | Authors: | Esposito, L, Bruno, I, Sica, F, Raia, C.A, Giordano, A, Rossi, M, Mazzarella, L, Zagari, A. | | Deposit date: | 2003-02-03 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural study of a single-point mutant of Sulfolobus solfataricus alcohol dehydrogenase with enhanced activity
Febs Lett., 539, 2003
|
|
1R37
 
 | | Alcohol dehydrogenase from sulfolobus solfataricus complexed with NAD(H) and 2-ethoxyethanol | | Descriptor: | 2-ETHOXYETHANOL, NAD-dependent alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Esposito, L, Bruno, I, Sica, F, Raia, C.A, Giordano, A, Rossi, M, Mazzarella, L, Zagari, A. | | Deposit date: | 2003-09-30 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a ternary complex of the alcohol dehydrogenase from Sulfolobus solfataricus
Biochemistry, 42, 2003
|
|
1NTO
 
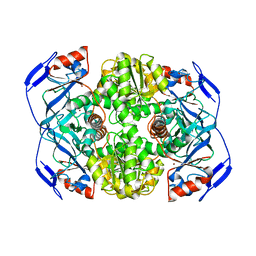 | | N249Y MUTANT OF ALCOHOL DEHYDROGENASE FROM THE ARCHAEON SULFOLOBUS SOLFATARICUS-MONOCLINIC CRYSTAL FORM | | Descriptor: | NAD-dependent alcohol dehydrogenase, ZINC ION | | Authors: | Esposito, L, Bruno, I, Sica, F, Raia, C.A, Giordano, A, Rossi, M, Mazzarella, L, Zagari, A. | | Deposit date: | 2003-01-30 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural study of a single-point mutant of Sulfolobus solfataricus alcohol dehydrogenase with enhanced activity.
Febs Lett., 539, 2003
|
|
1YV4
 
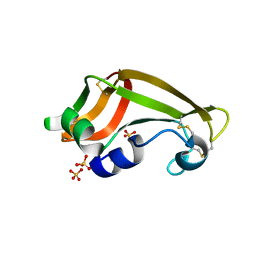 | | X-ray structure of M23L onconase at 100K | | Descriptor: | P-30 protein, SULFATE ION | | Authors: | Merlino, A, Mazzarella, L, Carannante, A, Di Fiore, A, Di Donato, A, Notomista, E, Sica, F. | | Deposit date: | 2005-02-15 | | Release date: | 2005-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The Importance of Dynamic Effects on the Enzyme Activity: X-RAY STRUCTURE AND MOLECULAR DYNAMICS OF ONCONASE MUTANTS
J.Biol.Chem., 280, 2005
|
|
1YV7
 
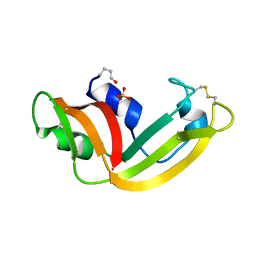 | | X-ray structure of (C87S,des103-104) onconase | | Descriptor: | P-30 protein, SULFATE ION | | Authors: | Merlino, A, Mazzarella, L, Carannante, A, Di Fiore, A, Di Donato, A, Notomista, E, Sica, F. | | Deposit date: | 2005-02-15 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Importance of Dynamic Effects on the Enzyme Activity: X-RAY STRUCTURE AND MOLECULAR DYNAMICS OF ONCONASE MUTANTS
J.Biol.Chem., 280, 2005
|
|
1YV6
 
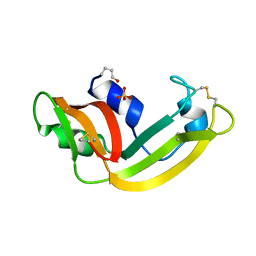 | | X-ray structure of M23L onconase at 298K | | Descriptor: | P-30 protein, SULFATE ION | | Authors: | Merlino, A, Mazzarella, L, Carannante, A, Di Fiore, A, Di Donato, A, Notomista, E, Sica, F. | | Deposit date: | 2005-02-15 | | Release date: | 2005-03-01 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Importance of Dynamic Effects on the Enzyme Activity: X-RAY STRUCTURE AND MOLECULAR DYNAMICS OF ONCONASE MUTANTS
J.Biol.Chem., 280, 2005
|
|
8RI5
 
 | | Crystal structure of transplatin/B-DNA adduct obtained upon 48 h of soaking | | Descriptor: | AMMONIA, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Tito, G, Troisi, R, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2023-12-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.415 Å) | | Cite: | On the mechanism of action of arsenoplatins: arsenoplatin-1 binding to a B-DNA dodecamer.
Dalton Trans, 53, 2024
|
|
8RI3
 
 | | Crystal structure of transplatin/B-DNA adduct obtained upon 7 days of soaking | | Descriptor: | AMMONIA, CHLORIDE ION, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), ... | | Authors: | Tito, G, Troisi, R, Ferraro, G, Sica, F, Merlino, A. | | Deposit date: | 2023-12-18 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | On the mechanism of action of arsenoplatins: arsenoplatin-1 binding to a B-DNA dodecamer.
Dalton Trans, 53, 2024
|
|
6EVV
 
 | | X-ray structure of the complex between human alpha thrombin and NU172, a duplex/quadruplex 26-mer DNA aptamer, in the presence of potassium ions. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Troisi, R, Russo Krauss, I, Sica, F. | | Deposit date: | 2017-11-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Several structural motifs cooperate in determining the highly effective anti-thrombin activity of NU172 aptamer.
Nucleic Acids Res., 46, 2018
|
|
6GN7
 
 | | X-ray structure of the complex between human alpha thrombin and NU172, a duplex/quadruplex 26-mer DNA aptamer, in the presence of sodium ions. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Troisi, R, Russo Krauss, I, Sica, F. | | Deposit date: | 2018-05-30 | | Release date: | 2018-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Several structural motifs cooperate in determining the highly effective anti-thrombin activity of NU172 aptamer.
Nucleic Acids Res., 46, 2018
|
|
1R3M
 
 | | Crystal structure of the dimeric unswapped form of bovine seminal ribonuclease | | Descriptor: | PHOSPHATE ION, Ribonuclease, seminal | | Authors: | Berisio, R, Sica, F, De Lorenzo, C, Di Fiore, A, Piccoli, R, Zagari, A, Mazzarella, L. | | Deposit date: | 2003-10-02 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the dimeric unswapped form of bovine seminal ribonuclease
Febs Lett., 554, 2003
|
|
5CMX
 
 | | X-ray structure of the complex between human alpha thrombin and a duplex/quadruplex 31-mer DNA aptamer | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, RE31, ... | | Authors: | Russo Krauss, I, Pica, A, Napolitano, V, Sica, F. | | Deposit date: | 2015-07-17 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Different duplex/quadruplex junctions determine the properties of anti-thrombin aptamers with mixed folding.
Nucleic Acids Res., 44, 2016
|
|
8BW5
 
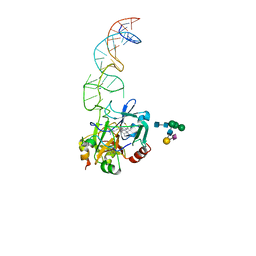 | | X-ray structure of the complex between human alpha thrombin and the duplex/quadruplex aptamer M08s-1_41mer | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, M08s-1_41mer, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Troisi, R, Napolitano, V, Sica, F. | | Deposit date: | 2022-12-06 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Steric hindrance and structural flexibility shape the functional properties of a guanine-rich oligonucleotide.
Nucleic Acids Res., 51, 2023
|
|
6Z8W
 
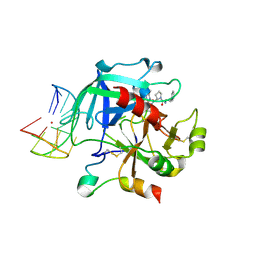 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3G), which contains 1-beta-D-glucopyranosyl residue in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
6Z8X
 
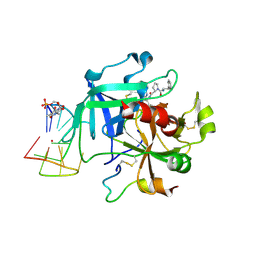 | | X-ray structure of the complex between human alpha thrombin and a thrombin binding aptamer variant (TBA-3Leu), which contains leucyl amide in the side chain of Thy3 at N3. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, Prothrombin, ... | | Authors: | Troisi, R, Timofeev, E.N, Sica, F. | | Deposit date: | 2020-06-02 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Expanding the recognition interface of the thrombin-binding aptamer HD1 through modification of residues T3 and T12.
Mol Ther Nucleic Acids, 23, 2021
|
|
