8ZJJ
 
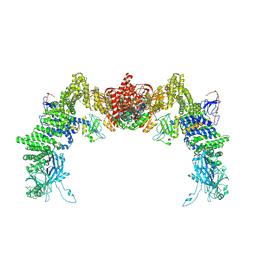 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 2) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJ2
 
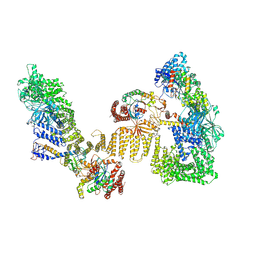 | | Cryo-EM structure of the RhoG/DOCK5/ELMO1/Rac1 complex | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJK
 
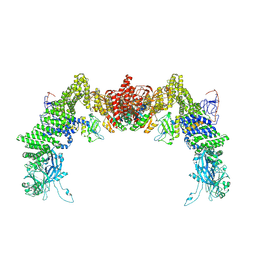 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 3) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJI
 
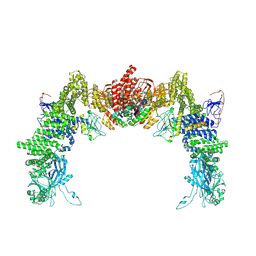 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 1) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8ZJM
 
 | | Structure of DOCK5/ELMO1/Rac1 core (RhoG/DOCK5/ELMO1/Rac1 dataset, class 5) | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.52 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
8JHK
 
 | | Cryo-EM structure of the DOCK5/ELMO1 complex, focused on one protomer | | Descriptor: | Dedicator of cytokinesis protein 5, Engulfment and cell motility protein 1 | | Authors: | Kukimoto-Niino, M, Katsura, K, Ishizuka-Katsura, Y, Mishima-Tsumagari, C, Yonemochi, M, Inoue, M, Nakagawa, R, Kaushik, R, Zhang, K.Y.J, Shirouzu, M. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.76 Å) | | Cite: | RhoG facilitates a conformational transition in the guanine nucleotide exchange factor complex DOCK5/ELMO1 to an open state.
J.Biol.Chem., 300, 2024
|
|
4URM
 
 | | Crystal Structure of Staph GyraseB 24kDa in complex with Kibdelomycin | | Descriptor: | (1R,4aS,5S,6S,8aR)-5-{[(5S)-1-(3-O-acetyl-4-O-carbamoyl-6-deoxy-2-O-methyl-alpha-L-talopyranosyl)-4-hydroxy-2-oxo-5-(propan-2-yl)-2,5-dihydro-1H-pyrrol-3-yl]carbonyl}-6-methyl-4-methylidene-1,2,3,4,4a,5,6,8a-octahydronaphthalen-1-yl 2,6-dideoxy-3-C-[(1S)-1-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}ethyl]-beta-D-ribo-hexopyranoside, DNA GYRASE SUBUNIT B | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-06-30 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
6B02
 
 | |
6B07
 
 | | Crystal structure of CfFPPS2, a lepidopteran type-II farnesyl diphosphate synthase, complexed with [1-phosphono-2-(1-propylpyridin-2-yl)ethyl]phosphonic acid (inhibitor 1d) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2,2-diphosphonoethyl)-1-propylpyridin-1-ium, Farnesyl diphosphate synthase, ... | | Authors: | Picard, M.-E, Cusson, M, Shi, R. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural characterization of a lepidopteran type-II farnesyl diphosphate synthase from the spruce budworm, Choristoneura fumiferana: Implications for inhibitor design.
Insect Biochem. Mol. Biol., 92, 2017
|
|
6B06
 
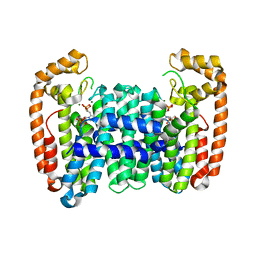 | | Crystal structure of CfFPPS2, a lepidopteran type-II farnesyl diphosphate synthase, complexed with IPP and [2-(1-methylpyridin-2-yl)-1-phosphono-ethyl]phosphonic acid (inhibitor 1b) | | Descriptor: | 2-(2,2-diphosphonoethyl)-1-methylpyridin-1-ium, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl diphosphate synthase, ... | | Authors: | Picard, M.-E, Cusson, M, Shi, R. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural characterization of a lepidopteran type-II farnesyl diphosphate synthase from the spruce budworm, Choristoneura fumiferana: Implications for inhibitor design.
Insect Biochem. Mol. Biol., 92, 2017
|
|
8K5W
 
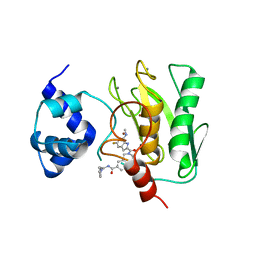 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | 2-[[5-fluoranyl-7-(methylamino)-1H-indol-2-yl]carbonyl]-N-(2-pyrrol-1-ylethyl)-3,4-dihydro-1H-isoquinoline-7-carboxamide, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
4MKP
 
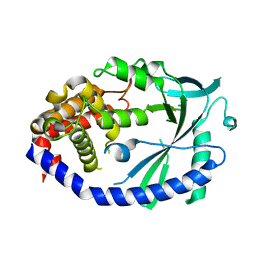 | | Crystal structure of human cGAS apo form | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2013-09-05 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Structural and Functional Analyses of DNA-Sensing and Immune Activation by Human cGAS
Plos One, 8, 2013
|
|
3TSP
 
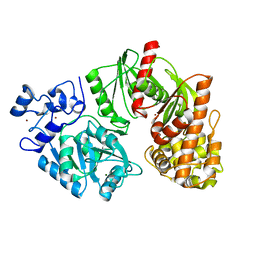 | | Crystal structure of E. coli HypF | | Descriptor: | MAGNESIUM ION, Transcriptional regulatory protein, ZINC ION | | Authors: | Petkun, S, Shi, R, Li, Y, Cygler, M. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Hydrogenase Maturation Protein HypF with Reaction Intermediates Shows Two Active Sites.
Structure, 19, 2011
|
|
3TSU
 
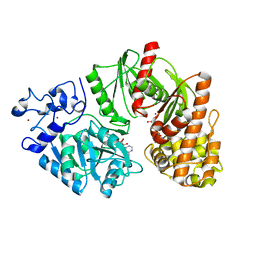 | | Crystal structure of E. coli HypF with AMP-PNP and carbamoyl phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Petkun, S, Shi, R, Li, Y, Cygler, M. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of Hydrogenase Maturation Protein HypF with Reaction Intermediates Shows Two Active Sites.
Structure, 19, 2011
|
|
3TTC
 
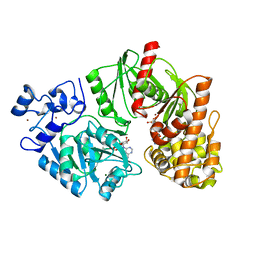 | | Crystal structure of E. coli HypF with ADP and carbamoyl phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transcriptional regulatory protein, ... | | Authors: | Petkun, S, Shi, R, Li, Y, Cygler, M. | | Deposit date: | 2011-09-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of Hydrogenase Maturation Protein HypF with Reaction Intermediates Shows Two Active Sites.
Structure, 19, 2011
|
|
3TTF
 
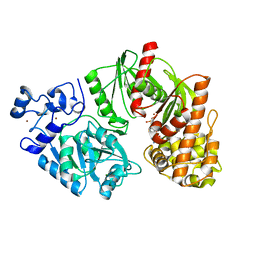 | | Crystal structure of E. coli HypF with AMP and carbamoyl phosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Transcriptional regulatory protein, ... | | Authors: | Petkun, S, Shi, R, Li, Y, Cygler, M. | | Deposit date: | 2011-09-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of Hydrogenase Maturation Protein HypF with Reaction Intermediates Shows Two Active Sites.
Structure, 19, 2011
|
|
3TSQ
 
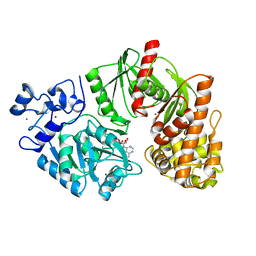 | | Crystal structure of E. coli HypF with ATP and Carbamoyl phosphate | | Descriptor: | 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, MAGNESIUM ION, Transcriptional regulatory protein, ... | | Authors: | Petkun, S, Shi, R, Li, Y, Cygler, M. | | Deposit date: | 2011-09-13 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Hydrogenase Maturation Protein HypF with Reaction Intermediates Shows Two Active Sites.
Structure, 19, 2011
|
|
3TTD
 
 | | Crystal structure of E. coli HypF with AMP-CPP and carbamoyl phosphate | | Descriptor: | MAGNESIUM ION, Transcriptional regulatory protein, ZINC ION | | Authors: | Petkun, S, Shi, R, Li, Y, Cygler, M. | | Deposit date: | 2011-09-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Hydrogenase Maturation Protein HypF with Reaction Intermediates Shows Two Active Sites.
Structure, 19, 2011
|
|
6LNG
 
 | | Rapid crystallization of streptavidin using charged peptides | | Descriptor: | GLYCEROL, Streptavidin | | Authors: | Minamihata, K, Tsukamoto, K, Adachi, M, Shimizu, R, Mishina, M, Kuroki, R, Nagamune, T. | | Deposit date: | 2019-12-30 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8000015 Å) | | Cite: | Genetically fused charged peptides induce rapid crystallization of proteins.
Chem.Commun.(Camb.), 56, 2020
|
|
5X2H
 
 | | Crystal structure of Campylobacter jejuni Cas9 in complex with sgRNA and target DNA (AGAAACA PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, Non-target DNA strand, ... | | Authors: | Yamada, M, Watanabe, Y, Hirano, H, Nakane, T, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems
Mol. Cell, 65, 2017
|
|
5X2G
 
 | | Crystal structure of Campylobacter jejuni Cas9 in complex with sgRNA and target DNA (AGAAACC PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, Non-target DNA strand, ... | | Authors: | Yamada, M, Watanabe, Y, Hirano, H, Nakane, T, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2017-01-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems
Mol. Cell, 65, 2017
|
|
6M04
 
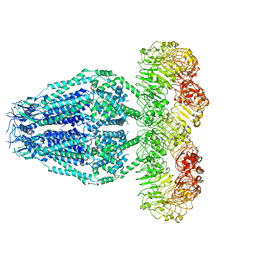 | | Structure of the human homo-hexameric LRRC8D channel at 4.36 Angstroms | | Descriptor: | Volume-regulated anion channel subunit LRRC8D | | Authors: | Nakamura, R, Kasuya, G, Yokoyama, T, Shirouzu, M, Ishitani, R, Nureki, O. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Cryo-EM structure of the volume-regulated anion channel LRRC8D isoform identifies features important for substrate permeation.
Commun Biol, 3, 2020
|
|
2PQ2
 
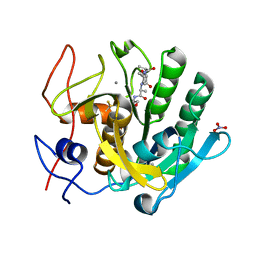 | | Structure of serine proteinase K complex with a highly flexible hydrophobic peptide at 1.8A resolution | | Descriptor: | CALCIUM ION, GALAG peptide, NITRATE ION, ... | | Authors: | Ethayathulla, A.S, Singh, A.K, Singh, N, Sharma, S, Sinha, M, Somvanshi, R.K, Kaur, P, Dey, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 2007-05-01 | | Release date: | 2007-05-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of serine proteinase K complex with a highly flexible hydrophobic peptide at 1.8A resolution
To be Published
|
|
4X5M
 
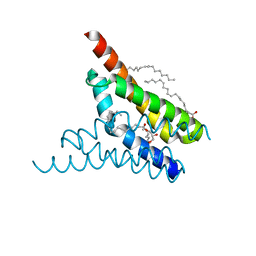 | | Crystal structure of SemiSWEET in the inward-open conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, OLEIC ACID, ... | | Authors: | Lee, Y, Nishizawa, T, Yamashita, K, Ishitani, R, Nureki, O. | | Deposit date: | 2014-12-05 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the facilitative diffusion mechanism by SemiSWEET transporter
Nat Commun, 6, 2015
|
|
1UG0
 
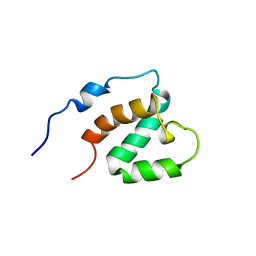 | | Solution structure of SURP domain in BAB30904 | | Descriptor: | splicing factor 4 | | Authors: | He, F, Muto, Y, Ushikoshi, R, Koshiba, S, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-11 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of SURP domain in BAB30904
To be Published
|
|
