7EK1
 
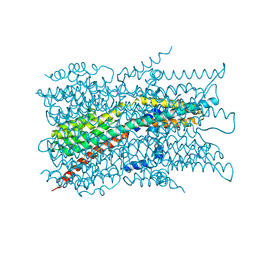 | | Cryo-EM structure of VCCN1 in detergent | | Descriptor: | Bestrophin-like protein | | Authors: | Hagino, T, Kato, T, Kasuya, G, Kobayashi, K, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of thylakoid-located voltage-dependent chloride channel VCCN1.
Nat Commun, 13, 2022
|
|
6K7N
 
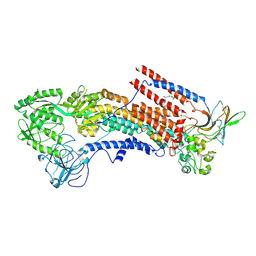 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1P state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
7EK3
 
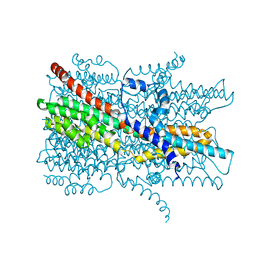 | | Cryo-EM structure of VCCN1 Y332A mutant in lipid nanodisc | | Descriptor: | Bestrophin-like protein | | Authors: | Hagino, T, Kato, T, Kasuya, G, Kobayashi, K, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of thylakoid-located voltage-dependent chloride channel VCCN1.
Nat Commun, 13, 2022
|
|
7V6B
 
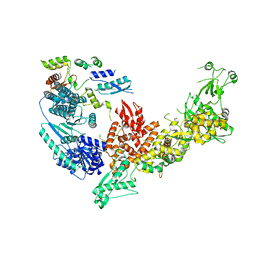 | | Structure of the Dicer-2-R2D2 heterodimer | | Descriptor: | Dicer-2, isoform A, R2D2 | | Authors: | Yamaguchi, S, Nishizawa, T, Kusakizako, T, Yamashita, K, Tomita, A, Hirano, H, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-20 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex.
Nature, 607, 2022
|
|
7V6C
 
 | | Structure of the Dicer-2-R2D2 heterodimer bound to small RNA duplex | | Descriptor: | Dicer-2, isoform A, R2D2, ... | | Authors: | Yamaguchi, S, Nishizawa, T, Kusakizako, T, Yamashita, K, Tomita, A, Hirano, H, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-20 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex.
Nature, 607, 2022
|
|
6K7G
 
 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1 state class1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
7FCI
 
 | | human NTCP in complex with YN69083 Fab | | Descriptor: | Fab Heavy chain, Fab Light chain, Sodium/bile acid cotransporter | | Authors: | Park, J.H, Iwamoto, M, Yun, J.H, Uchikubo-Kamo, T, Son, D, Jin, Z, Yoshida, H, Ohki, M, Ishimoto, N, Mizutani, K, Oshima, M, Muramatsu, M, Wakita, T, Shirouzu, M, Liu, K, Uemura, T, Nomura, N, Iwata, S, Watashi, K, Tame, J.R.H, Nishizawa, T, Lee, W, Park, S.Y. | | Deposit date: | 2021-07-14 | | Release date: | 2022-05-25 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the HBV receptor and bile acid transporter NTCP.
Nature, 606, 2022
|
|
7V94
 
 | | Cryo-EM structure of the Cas12c2-sgRNA-target DNA ternary complex | | Descriptor: | Cas12c2, sgRNA, target DNA (non target strand), ... | | Authors: | Kurihara, N, Hirano, H, Tomita, A, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the type V-C CRISPR-Cas effector enzyme.
Mol.Cell, 82, 2022
|
|
7V93
 
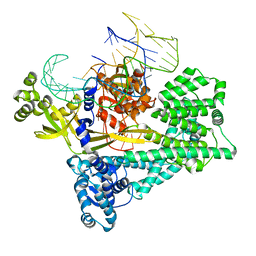 | | Cryo-EM structure of the Cas12c2-sgRNA binary complex | | Descriptor: | cas12c2, sgRNA | | Authors: | Kurihara, N, Hirano, H, Tomita, A, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-08-24 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the type V-C CRISPR-Cas effector enzyme.
Mol.Cell, 82, 2022
|
|
6K7L
 
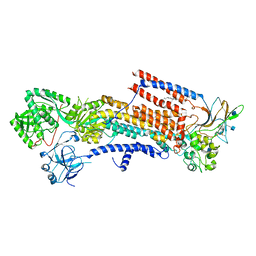 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E2P state class2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
6K7M
 
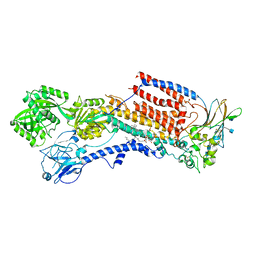 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E2Pi-PL state) | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-3-hexadecanoyloxy-2-[(~{Z})-octadec-9-enoyl]oxy-propoxy]-oxidanyl-phosphoryl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
7DB6
 
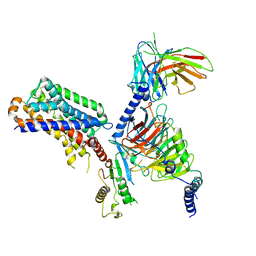 | | human melatonin receptor MT1 - Gi1 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Okamoto, H.H, Kusakizako, T, Shihioya, W, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-10-19 | | Release date: | 2021-08-18 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the human MT 1 -G i signaling complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6K7K
 
 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1-ADP-Pi state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
6K7J
 
 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1-ATP state class1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
7E8D
 
 | | NSD2 E1099K mutant bound to nucleosome | | Descriptor: | DNA (185-MER), Histone H2A type 1, Histone H2B type 1-J, ... | | Authors: | Sengoku, T, Sato, K, Nishizawa, T, Nureki, O, Ogata, K. | | Deposit date: | 2021-03-01 | | Release date: | 2021-11-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of the regulation of the normal and oncogenic methylation of nucleosomal histone H3 Lys36 by NSD2.
Nat Commun, 12, 2021
|
|
7C7L
 
 | | Cryo-EM structure of the Cas12f1-sgRNA-target DNA complex | | Descriptor: | CRISPR-associated protein Cas14a.1, DNA (40-mer), ZINC ION, ... | | Authors: | Takeda, N.S, Nakagawa, R, Okazaki, S, Hirano, H, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2020-05-26 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the miniature type V-F CRISPR-Cas effector enzyme.
Mol.Cell, 81, 2021
|
|
7WAH
 
 | | Structure of Cas7-11 in complex with guide RNA and target RNA | | Descriptor: | CRISPR-associated RAMP family protein, ZINC ION, crRNA (39-MER), ... | | Authors: | Kato, K, Okazaki, S, Isayama, Y, Nishizawa, T, Nishimasu, H. | | Deposit date: | 2021-12-14 | | Release date: | 2022-06-15 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structure and engineering of the type III-E CRISPR-Cas7-11 effector complex.
Cell, 185, 2022
|
|
7CCS
 
 | | Consensus mutated xCT-CD98hc complex | | Descriptor: | 4F2 cell-surface antigen heavy chain, Consensus mutated Anionic Amino Acid Transporter Light Chain, Xc- System | | Authors: | Oda, K, Lee, Y, Takemoto, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-06-17 | | Release date: | 2020-12-09 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Consensus mutagenesis approach improves the thermal stability of system x c - transporter, xCT, and enables cryo-EM analyses.
Protein Sci., 29, 2020
|
|
7F47
 
 | | Cryo-EM structure of Rhizobium etli MprF | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Hypothetical conserved protein, [(2R)-1-[[(2R)-3-[(2S)-2,6-bis(azanyl)hexanoyl]oxy-2-oxidanyl-propoxy]-oxidanyl-phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (E)-octadec-9-enoate | | Authors: | Nishimura, M, Hirano, H, Kobayashi, K, Gill, C.P, Phan, C.N.K, Kise, Y, Kusakizako, T, Yamashita, K, Ito, Y, Roy, H, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-06-17 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structure of Rhizobium etli MprF
To Be Published
|
|
7EFC
 
 | | 1.70 A cryo-EM structure of streptavidin | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Kotecha, A, Nureki, O. | | Deposit date: | 2021-03-21 | | Release date: | 2021-04-28 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.7 Å) | | Cite: | 1.70 A cryo-EM structure of streptavidin using all frames (corresponding to 70 e/A^2 total dose)
To Be Published
|
|
7EFD
 
 | | 1.77 A cryo-EM structure of Streptavidin using first 40 frames (corresponding to about 40 e/A^2 total dose) | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Kotecha, A, Nureki, O. | | Deposit date: | 2021-03-21 | | Release date: | 2021-04-28 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.77 Å) | | Cite: | 1.77 A cryo-EM structure of Streptavidin using first 40 frames (corresponding to about 40e/A^2 total dose)
To Be Published
|
|
7DY0
 
 | |
