6ZWT
 
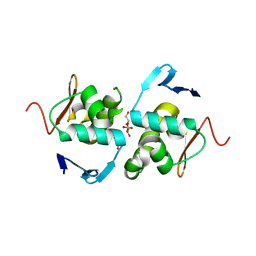 | | Crystal structure of DNA-binding domain of OmpR of two-component system of Acinetobacter baumannii | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Narwal, M, Lucchini, V, Trebosc, V, Gartenmann, S, Pieren, M, Kemmer, C, Kammerer, R.A. | | Deposit date: | 2020-07-28 | | Release date: | 2021-08-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting virulence regulation to disarm Acinetobacter baumannii pathogenesis.
Virulence, 13, 2022
|
|
4UVS
 
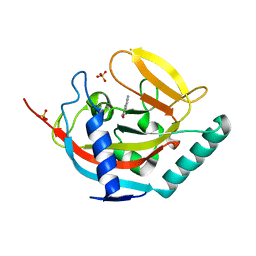 | | Crystal structure of human tankyrase 2 in complex with 5-amino-3- pentyl-1,2-dihydroisoquinolin-1-one | | Descriptor: | 5-amino-3-pentylisoquinolin-1(2H)-one, SULFATE ION, TANKYRASE-2, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4UVP
 
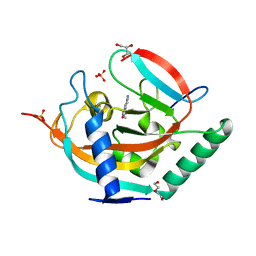 | | Crystal structure of human tankyrase 2 in complex with 5-amino-3- ethyl-1,2-dihydroisoquinolin-1-one | | Descriptor: | 5-amino-3-ethylisoquinolin-1(2H)-one, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-07 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4ZTB
 
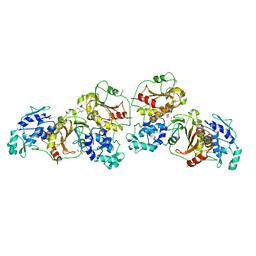 | | Crystal structure of nsP2 protease from Chikungunya virus in P212121 space group at 2.59 A (4molecules/ASU). | | Descriptor: | GLYCEROL, Protease nsP2 | | Authors: | Narwal, M, Pratap, S, Singh, H, Kumar, P, Tomar, S. | | Deposit date: | 2015-05-14 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of chikungunya virus nsP2 cysteine protease reveals a putative flexible loop blocking its active site.
Int.J.Biol.Macromol., 116, 2018
|
|
4UVN
 
 | | Crystal structure of human tankyrase 2 in complex with 5-amino-3-(4- chlorophenyl)-1,2-dihydroisoquinolin-1-one | | Descriptor: | 5-amino-3-(4-chlorophenyl)isoquinolin-1(2H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-07 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
8EIR
 
 | | SARS-CoV-2 polyprotein substrate regulates the stepwise Mpro cleavage reaction | | Descriptor: | 3C-like proteinase nsp5, nsp7-nsp10 of Replicase polyprotein 1a | | Authors: | Narwal, M, Edwards, T, Armache, J.P, Murakami, K.S. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | SARS-CoV-2 polyprotein substrate regulates the stepwise M pro cleavage reaction.
J.Biol.Chem., 299, 2023
|
|
8EKE
 
 | |
4UVZ
 
 | | Crystal structure of human tankyrase 2 in complex with 5-amino-3- phenyl-1,2-dihydroisoquinolin-1-one | | Descriptor: | 5-AMINO-3-PHENYL-1,2-DIHYDROISOQUINOLIN-1-ONE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4UVO
 
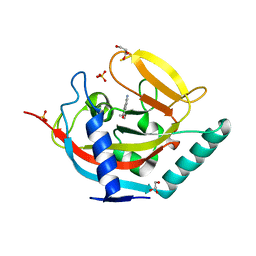 | | Crystal structure of human tankyrase 2 in complex with 5-amino-3-(4- methoxyphenyl)-1,2-dihydroisoquinolin-1-one | | Descriptor: | 5-amino-3-(4-methoxyphenyl)isoquinolin-1(2H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-07 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4UVL
 
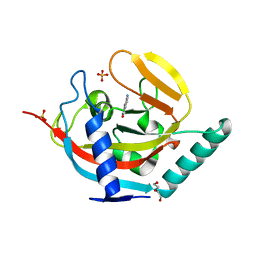 | | Crystal structure of human tankyrase 2 in complex with 5-amino-1,2- dihydroisoquinolin-1-one | | Descriptor: | 5-aminoisoquinolin-1(4H)-one, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-07 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4UVT
 
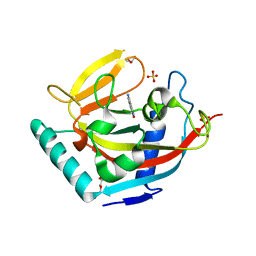 | | Crystal structure of human tankyrase 2 in complex with 5-amino-4- methyl-1,2-dihydroisoquinolin-1-one | | Descriptor: | 5-amino-4-methylisoquinolin-1(2H)-one, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
5MZF
 
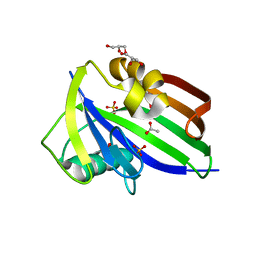 | | Crystal structure of dog MTH1 protein | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Narwal, M, Jemth, A.-S, Helleday, T, Stenmark, P. | | Deposit date: | 2017-01-31 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures and Inhibitor Interactions of Mouse and Dog MTH1 Reveal Species-Specific Differences in Affinity.
Biochemistry, 57, 2018
|
|
5MZG
 
 | | Crystal structure of mouse MTH1 in complex with TH588 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, N~4~-cyclopropyl-6-(2,3-dichlorophenyl)pyrimidine-2,4-diamine, SULFATE ION, ... | | Authors: | Narwal, M, Jemth, A.-S, Helleday, T, Stenmark, P. | | Deposit date: | 2017-01-31 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures and Inhibitor Interactions of Mouse and Dog MTH1 Reveal Species-Specific Differences in Affinity.
Biochemistry, 57, 2018
|
|
5MZE
 
 | | Crystal structure of mouse MTH1 with 8-oxo-dGTP | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, COPPER (II) ION, ... | | Authors: | Narwal, M, Jemth, A.-S, Helleday, T, Stenmark, P. | | Deposit date: | 2017-01-31 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures and Inhibitor Interactions of Mouse and Dog MTH1 Reveal Species-Specific Differences in Affinity.
Biochemistry, 57, 2018
|
|
5HZX
 
 | | Crystal structure of zebrafish MTH1 in complex with TH588 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Narwal, M, Gustafsson, R, Brautigam, L, Pudelko, L, Jemth, A.-S, Gad, H, Karsten, S, Carreras-Puigvert, J, Homan, E, Berndt, C, Berglund, U.W, Helleday, T, Stenmark, P. | | Deposit date: | 2016-02-03 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hypoxic Signaling and the Cellular Redox Tumor Environment Determine Sensitivity to MTH1 Inhibition.
Cancer Res., 76, 2016
|
|
3UA9
 
 | | Crystal structure of human tankyrase 2 in complex with a selective inhibitor | | Descriptor: | 4-[(3aR,4S,7R,7aS)-1,3-dioxo-1,3,3a,4,7,7a-hexahydro-2H-4,7-methanoisoindol-2-yl]-N-(quinolin-8-yl)benzamide, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Narwal, M, Lehtio, L. | | Deposit date: | 2011-10-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of selective inhibition of human tankyrases.
J.Med.Chem., 55, 2012
|
|
7LKE
 
 | |
7LKD
 
 | |
3U9H
 
 | |
3U9Y
 
 | |
7LBN
 
 | |
4HLM
 
 | | Crystal structure of Tankyrase 2 in complex with 3',4'-Dihydroxyflavone | | Descriptor: | 2-(3,4-dihydroxyphenyl)-4H-chromen-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2012-10-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Screening and structural analysis of flavones inhibiting tankyrases.
J.Med.Chem., 56, 2013
|
|
4HMH
 
 | | Crystal structure of tankyrase 2 in complex with 7,3-dihydroxyflavone | | Descriptor: | 7-hydroxy-2-(3-hydroxyphenyl)-4H-chromen-4-one, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2012-10-18 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Screening and structural analysis of flavones inhibiting tankyrases.
J.Med.Chem., 56, 2013
|
|
4HLF
 
 | | Crystal structure of Tankyrase 2 in complex with 7,3',4'-Trihydroxyflavone | | Descriptor: | 2-(3,4-dihydroxyphenyl)-7-hydroxy-4H-chromen-4-one, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2012-10-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Screening and structural analysis of flavones inhibiting tankyrases.
J.Med.Chem., 56, 2013
|
|
4HLG
 
 | | Crystal structure of Tankyrase 2 in complex with 3'-hydroxyflavone | | Descriptor: | 2-(3-hydroxyphenyl)-4H-chromen-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2012-10-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Screening and structural analysis of flavones inhibiting tankyrases.
J.Med.Chem., 56, 2013
|
|
