8FM9
 
 | | Nodavirus RNA replication proto-crown, detergent-solubliized C12 multimer | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Zhan, H, Unchwaniwala, N, Rebolledo Viveros, A, Pennington, J, Horswill, M, Broadberry, R, Myers, J, den Boon, J, Grant, T, Ahlquist, P. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FHD
 
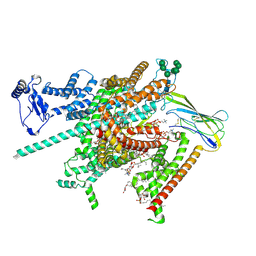 | | Cryo-EM structure of human voltage-gated sodium channel Nav1.6 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, (5E,17R,20S)-23-amino-20-hydroxy-14,20-dioxo-15,19,21-trioxa-20lambda~5~-phosphatricos-5-en-17-yl hexadecanoate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2022-12-14 | | Release date: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of human voltage-gated sodium channel Na v 1.6.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3PCQ
 
 | | Femtosecond X-ray protein Nanocrystallography | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Chapman, H.N, Fromme, P, Barty, A, White, T.A, Kirian, R.A, Aquila, A, Hunter, M.S, Schulz, J, Deponte, D.P, Weierstall, U, Doak, R.B, Maia, F.R.N.C, Martin, A.V, Schlichting, I, Lomb, L, Coppola, N, Shoeman, R.L, Epp, S.W, Hartmann, R, Rolles, D, Rudenko, A, Foucar, L, Kimmel, N, Weidenspointner, G, Holl, P, Liang, M, Barthelmess, M, Caleman, C, Boutet, S, Bogan, M.J, Krzywinski, J, Bostedt, C, Bajt, S, Gumprecht, L, Rudek, B, Erk, B, Schmidt, C, Homke, A, Reich, C, Pietschner, D, Struder, L, Hauser, G, Gorke, H, Ullrich, J, Herrmann, S, Schaller, G, Schopper, F, Soltau, H, Kuhnel, K.-U, Messerschmidt, M, Bozek, J.D, Hau-Riege, S.P, Frank, M, Hampton, C.Y, Sierra, R, Starodub, D, Williams, G.J, Hajdu, J, Timneanu, N, Seibert, M.M, Andreasson, J, Rocker, A, Jonsson, O, Svenda, M, Stern, S, Nass, K, Andritschke, R, Schroter, C.-D, Krasniqi, F, Bott, M, Schmidt, K.E, Wang, X, Grotjohann, I, Holton, J.M, Barends, T.R.M, Neutze, R, Marchesini, S, Fromme, R, Schorb, S, Rupp, D, Adolph, M, Gorkhover, T, Andersson, I, Hirsemann, H, Potdevin, G, Graafsma, H, Nilsson, B, Spence, J.C.H. | | Deposit date: | 2010-10-21 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (8.984 Å) | | Cite: | Femtosecond X-ray protein nanocrystallography.
Nature, 470, 2011
|
|
6W4M
 
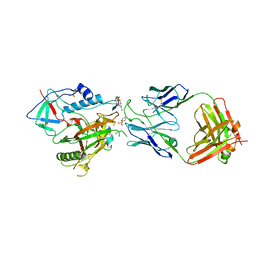 | | CRYSTAL STRUCTURE OF THE ADCC-POTENT, WEAKLY NEUTRALIZING HIV ENV CO-RECEPTOR BINDING SITE ANTIBODY N12-I2 FAB IN COMPLEX WITH HIV-1 CLADE A/E GP120 AND M48U1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTI-HIV ANTIBODY N12-I2 FAB HEAVY CHAIN, ANTI-HIV ANTIBODY N12-I2 FAB LIGHT CHAIN, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2020-03-11 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Defining rules governing recognition and Fc-mediated effector functions to the HIV-1 co-receptor binding site.
Bmc Biol., 18, 2020
|
|
1BM1
 
 | | CRYSTAL STRUCTURE OF BACTERIORHODOPSIN IN THE LIGHT-ADAPTED STATE | | Descriptor: | BACTERIORHODOPSIN, PHOSPHORIC ACID 2,3-BIS-(3,7,11,15-TETRAMETHYL-HEXADECYLOXY)-PROPYL ESTER 2-HYDROXO-3-PHOSPHONOOXY-PROPYL ESTER, RETINAL | | Authors: | Sato, H, Takeda, K, Tani, K, Hino, T, Okada, T, Nakasako, M, Kamiya, N, Kouyama, T. | | Deposit date: | 1998-07-28 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Specific lipid-protein interactions in a novel honeycomb lattice structure of bacteriorhodopsin.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
8FMB
 
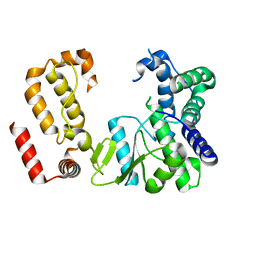 | | Nodavirus RNA replication protein A polymerase domain, local refinement | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Zhan, H, Unchwaniwala, N, Rebolledo Viveros, A, Pennington, J, Horswill, M, Broadberry, R, Myers, J, den Boon, J, Grant, T, Ahlquist, P. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6VXO
 
 | | NaChBac-Nav1.7VSDII chimera in nanodisc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, NaChBac-Nav1.7VSDII chimera | | Authors: | Yan, N, Gao, S. | | Deposit date: | 2020-02-22 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Employing NaChBac for cryo-EM analysis of toxin action on voltage-gated Na+channels in nanodisc.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1UE5
 
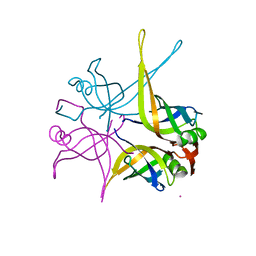 | | Crystal structure of the single-stranded dna-binding protein from mycobacterium tuberculosis | | Descriptor: | CADMIUM ION, Single-strand binding protein | | Authors: | Saikrishnan, K, Jeyakanthan, J, Venkatesh, J, Acharya, N, Sekar, K, Varshney, U, Vijayan, M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-05-09 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Mycobacterium tuberculosis single-stranded DNA-binding protein. Variability in quaternary structure and its implications
J.MOL.BIOL., 331, 2003
|
|
1BXY
 
 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L30 FROM THERMUS THERMOPHILUS AT 1.9 A RESOLUTION: CONFORMATIONAL FLEXIBILITY OF THE MOLECULE. | | Descriptor: | PROTEIN (RIBOSOMAL PROTEIN L30) | | Authors: | Fedorov, R, Nevskaya, N, Khairullina, A, Tishchenko, S, Mikhailov, A, Garber, M, Nikonov, S. | | Deposit date: | 1998-10-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of ribosomal protein L30 from Thermus thermophilus at 1.9 A resolution: conformational flexibility of the molecule.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BK0
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ACV-FE COMPLEX) | | Descriptor: | FE (III) ION, ISOPENICILLIN N SYNTHASE, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, ... | | Authors: | Roach, P.L, Clifton, I.J, Hensgens, C.M.H, Shibata, N, Schofield, C.J, Hajdu, J, Baldwin, J.E. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation.
Nature, 387, 1997
|
|
7QBS
 
 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, S-adenosyl-L-methionine, and peptide bound. | | Descriptor: | CO-METHYLCOBALAMIN, FE (III) ION, IRON/SULFUR CLUSTER, ... | | Authors: | Bernardo-Garcia, N, Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
7QGI
 
 | | Crystal structure of SARS-CoV-2 NSP14 in the absence of NSP10 | | Descriptor: | PHOSPHATE ION, Proofreading exoribonuclease nsp14, ZINC ION | | Authors: | Newman, J.A, Imprachim, N, Yosaatmadja, Y, Gileadi, O. | | Deposit date: | 2021-12-08 | | Release date: | 2022-01-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures and fragment screening of SARS-CoV-2 NSP14 reveal details of exoribonuclease activation and mRNA capping and provide starting points for antiviral drug development.
Nucleic Acids Res., 51, 2023
|
|
1BTP
 
 | | UNIQUE BINDING OF A NOVEL SYNTHETIC INHIBITOR, N-[3-[4-[4-(AMIDINOPHENOXY)-CARBONYL]PHENYL]-2-METHYL-2-PROPENOYL]-N-ALLYLGLYCINE METHANESULFONATE TO BOVINE TRYPSIN, REVEALED BY THE CRYSTAL STRUCTURE OF THE COMPLEX | | Descriptor: | BETA-TRYPSIN, CALCIUM ION | | Authors: | Odagaki, Y, Nakai, H, Senokuchi, K, Kawamura, M, Hamanaka, N, Nakamura, M, Tomoo, K, Ishida, T. | | Deposit date: | 1995-08-11 | | Release date: | 1996-01-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique binding of a novel synthetic inhibitor, N-[3-[4-[4-(amidinophenoxy)carbonyl]phenyl]-2-methyl-2-propenoyl]- N-allylglycine methanesulfonate, to bovine trypsin, revealed by the crystal structure of the complex.
Biochemistry, 34, 1995
|
|
6VME
 
 | | Human ESCRT-I heterotetramer headpiece | | Descriptor: | Multivesicular body subunit 12A, Tumor susceptibility gene 101 protein, Vacuolar protein sorting-associated protein 28 homolog, ... | | Authors: | Flower, T.G, Hurley, J.H, Tjahjono, N. | | Deposit date: | 2020-01-27 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | A helical assembly of human ESCRT-I scaffolds reverse-topology membrane scission.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7QIF
 
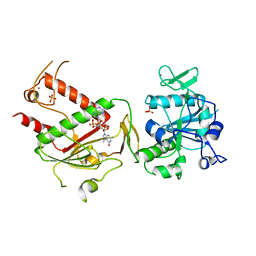 | | Crystal structure of SARS-CoV-2 NSP14 in complex with 7MeGpppG. | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Newman, J.A, Imprachim, N, Yosaatmadja, Y, Gileadi, O. | | Deposit date: | 2021-12-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structures and fragment screening of SARS-CoV-2 NSP14 reveal details of exoribonuclease activation and mRNA capping and provide starting points for antiviral drug development.
Nucleic Acids Res., 51, 2023
|
|
1U6T
 
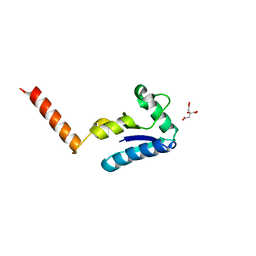 | | Crystal structure of the human SH3 binding glutamic-rich protein like | | Descriptor: | CITRIC ACID, SH3 domain-binding glutamic acid-rich-like protein | | Authors: | Yin, L, Xiang, Y, Yang, N, Zhu, D.-Y, Huang, R.-H, Wang, D.-C. | | Deposit date: | 2004-08-01 | | Release date: | 2005-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human SH3BGRL protein: the first structure of the human SH3BGR family representing a novel class of thioredoxin fold proteins
Proteins, 61, 2005
|
|
3A9E
 
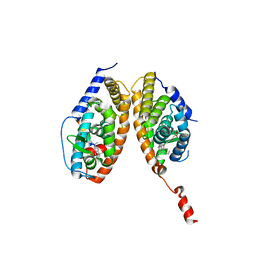 | | Crystal structure of a mixed agonist-bound RAR-alpha and antagonist-bound RXR-alpha heterodimer ligand binding domains | | Descriptor: | (2E,4E,6Z)-3-methyl-7-(5,5,8,8-tetramethyl-3-propoxy-5,6,7,8-tetrahydronaphthalen-2-yl)octa-2,4,6-trienoic acid, 13-mer (LXXLL motif) from Nuclear receptor coactivator 2, RETINOIC ACID, ... | | Authors: | Sato, Y, Duclaud, S, Peluso-Iltis, C, Poussin, P, Moras, D, Rochel, N, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2009-10-24 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Phantom Effect of the Rexinoid LG100754: structural and functional insights
Plos One, 5, 2010
|
|
1C3L
 
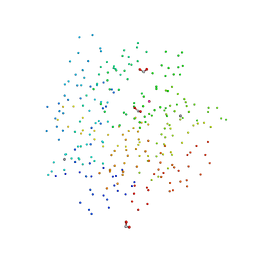 | | SUBTILISIN-CARLSBERG COMPLEXED WITH XENON (8 BAR) | | Descriptor: | CALCIUM ION, FORMIC ACID, SUBTILISIN-CARLSBERG, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S. | | Deposit date: | 1999-07-28 | | Release date: | 1999-08-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
1BMR
 
 | | ALPHA-LIKE TOXIN LQH III FROM SCORPION LEIURUS QUINQUESTRIATUS HEBRAEUS, NMR, 25 STRUCTURES | | Descriptor: | LQH III ALPHA-LIKE TOXIN | | Authors: | Krimm, I, Gilles, N, Sautiere, P, Stankiewicz, M, Pelhate, M, Gordon, D, Lancelin, J.-M. | | Deposit date: | 1998-07-24 | | Release date: | 1999-02-16 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | NMR structures and activity of a novel alpha-like toxin from the scorpion Leiurus quinquestriatus hebraeus.
J.Mol.Biol., 285, 1999
|
|
6VWX
 
 | | NaChBac in lipid nanodisc | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, BH1501 protein, SODIUM ION | | Authors: | Yan, N, Gao, S. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Employing NaChBac for cryo-EM analysis of toxin action on voltage-gated Na+channels in nanodisc.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VX9
 
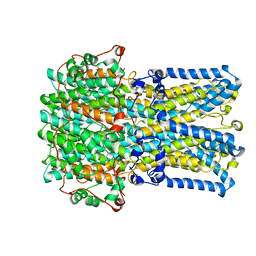 | | bestrophin-2 Ca2+- unbound state 1 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7QGM
 
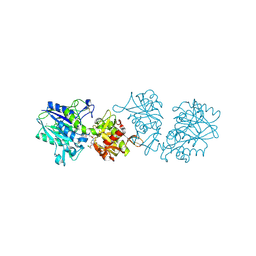 | |
7QGL
 
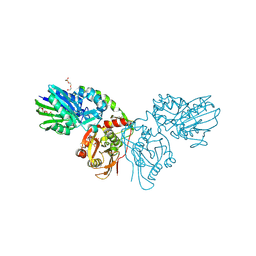 | |
6VX8
 
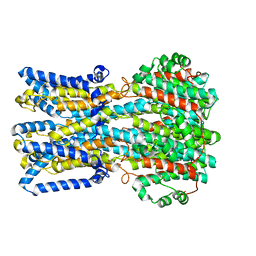 | | bestrophin-2 Ca2+- unbound state 2 (EGTA only) | | Descriptor: | Bestrophin, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7QGO
 
 | | Human CD73 (ecto 5'-nucleotidase) in complex with MRS4602 (a 3-methyl-CMPCP derivative, compound 21 in paper) in the closed state (crystal form III) | | Descriptor: | 5'-nucleotidase, ZINC ION, [[(2~{R},3~{S},4~{R},5~{R})-5-[(4~{E})-4-[(4-methoxycarbonylphenyl)methoxyimino]-3-methyl-2-oxidanylidene-pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]methylphosphonic acid | | Authors: | Strater, N. | | Deposit date: | 2021-12-09 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structure-Activity Relationship of 3-Methylcytidine-5'-alpha , beta-methylenediphosphates as CD73 Inhibitors.
J.Med.Chem., 65, 2022
|
|
