6DS5
 
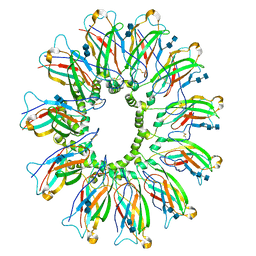 | | Cryo EM structure of human SEIPIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Seipin | | Authors: | Yan, R.H, Qian, H.W, Yan, N, Yang, H.Y. | | Deposit date: | 2018-06-13 | | Release date: | 2018-10-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Human SEIPIN Binds Anionic Phospholipids.
Dev. Cell, 47, 2018
|
|
6QCK
 
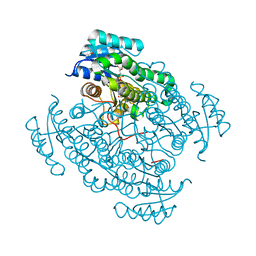 | | 17beta-hydroxysteroid dehydrogenase 14 variant T205 in complex with FB262 | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, 2-[2-(1,3-benzodioxol-2-yl)ethyl]benzoic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Bertoletti, N, Marchais-Oberwinkler, S, Heine, A, Klebe, G. | | Deposit date: | 2018-12-28 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Mutational and structural studies uncover crucial amino acids determining activity and stability of 17 beta-HSD14.
J.Steroid Biochem.Mol.Biol., 189, 2019
|
|
6DQJ
 
 | | Human type 3 1,4,5-inositol trisphosphate receptor in a ligand-free state | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DSO
 
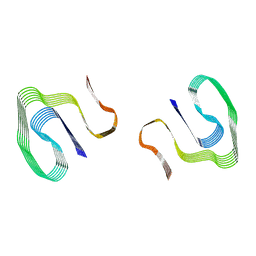 | | Cryo-EM structure of murine AA amyloid fibril | | Descriptor: | Serum amyloid A-2 protein | | Authors: | Loerch, S, Liberta, F, Grigorieff, N, Fandrich, M, Schmidt, M. | | Deposit date: | 2018-06-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM fibril structures from systemic AA amyloidosis reveal the species complementarity of pathological amyloids.
Nat Commun, 10, 2019
|
|
6DSV
 
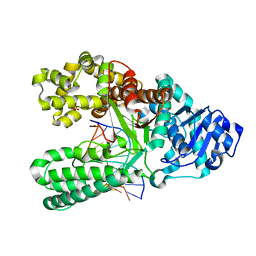 | | Bst DNA polymerase I post-chemistry (n+2) structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
6QD1
 
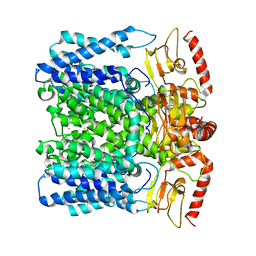 | | MloK1 model from single particle analysis of 2D crystals, class 5 (intermediate compact conformation) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Righetto, R, Biyani, N, Kowal, J, Chami, M, Stahlberg, H. | | Deposit date: | 2018-12-31 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM.
Nat Commun, 10, 2019
|
|
6UD0
 
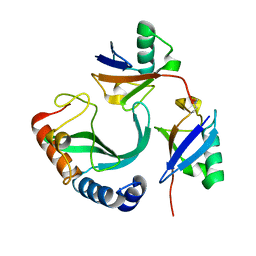 | | Solution-state NMR structural ensemble of human Tsg101 UEV in complex with K63-linked diubiquitin | | Descriptor: | Tumor susceptibility gene 101 protein, Ubiquitin | | Authors: | Strickland, M, Watanabe, S, Bonn, S.M, Camara, C.M, Fushman, D, Carter, C.A, Tjandra, N. | | Deposit date: | 2019-09-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Tsg101/ESCRT-I Recruitment Regulated by the Dual Binding Modes of K63-Linked Diubiquitin
Structure, 2021
|
|
5EDM
 
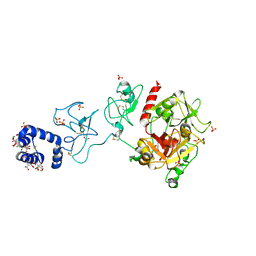 | | Crystal structure of prothrombin deletion mutant residues 154-167 ( Form I ) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Pozzi, N, Chen, Z, Di Cera, E. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How the Linker Connecting the Two Kringles Influences Activation and Conformational Plasticity of Prothrombin.
J.Biol.Chem., 291, 2016
|
|
5DO0
 
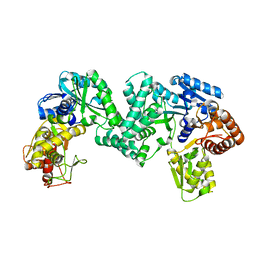 | | The structure of PKMT1 from Rickettsia prowazekii | | Descriptor: | protein lysine methyltransferase 1 | | Authors: | Noinaj, N, Abeykoon, A, He, Y, Yang, D.C, Buchanan, S.K. | | Deposit date: | 2015-09-10 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into Substrate Recognition and Catalysis in Outer Membrane Protein B (OmpB) by Protein-lysine Methyltransferases from Rickettsia.
J.Biol.Chem., 291, 2016
|
|
6DWJ
 
 | | SAMHD1 Bound to Vidarabine-TP in the Catalytic Pocket | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 9-{5-O-[(S)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}-9H-purin-6-amine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Knecht, K.M, Buzovetsky, O, Schneider, C, Thomas, D, Srikanth, V, Kaderali, L, Tofoleanu, F, Reiss, K, Ferreiros, N, Geisslinger, G, Batista, V.S, Ji, X, Cinatl, J, Keppler, O.T, Xiong, Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6UNQ
 
 | | Kinase domain of ALK2-K493A with AMPPNP | | Descriptor: | Activin receptor type-1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Agnew, C, Jura, N. | | Deposit date: | 2019-10-13 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Nat Commun, 12, 2021
|
|
6UNR
 
 | | Kinase domain of ALK2-K492A/K493A with AMPPNP | | Descriptor: | Activin receptor type-1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Agnew, C, Jura, N. | | Deposit date: | 2019-10-13 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Nat Commun, 12, 2021
|
|
6UNP
 
 | | Crystal structure of the kinase domain of BMPR2-D485G | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, Bone morphogenetic protein receptor type-2, ... | | Authors: | Agnew, C, Jura, N. | | Deposit date: | 2019-10-13 | | Release date: | 2021-07-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Nat Commun, 12, 2021
|
|
6UNS
 
 | | Kinase domain of ALK2-K492A/K493A with LDN-193189 | | Descriptor: | 4-[6-(4-piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline, Activin receptor type-1 | | Authors: | Agnew, C, Jura, N. | | Deposit date: | 2019-10-13 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for ALK2/BMPR2 receptor complex signaling through kinase domain oligomerization.
Nat Commun, 12, 2021
|
|
1OVL
 
 | | Crystal Structure of Nurr1 LBD | | Descriptor: | BROMIDE ION, IODIDE ION, Orphan nuclear receptor NURR1 (MSE 414, ... | | Authors: | Wang, Z, Liu, J, Walker, N. | | Deposit date: | 2003-03-26 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Function of Nurr1 identifies a Class of Ligand-Independent Nuclear Receptors
Nature, 423, 2003
|
|
6DEU
 
 | | Human caspase-6 A109T | | Descriptor: | Caspase-6 | | Authors: | Tubeleviciute-Aydin, A, Beautrait, A, Lynham, J, Sharma, G, Gorelik, A, Deny, L.J, Soya, N, Lukacs, G.L, Nagar, B, Marinier, A, LeBlanc, A.C. | | Deposit date: | 2018-05-13 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Identification of Allosteric Inhibitors against Active Caspase-6.
Sci Rep, 9, 2019
|
|
1B0Q
 
 | | DITHIOL ALPHA MELANOTROPIN PEPTIDE CYCLIZED VIA RHENIUM METAL COORDINATION | | Descriptor: | PROTEIN (CYCLIC ALPHA MELANOCYTE STIMULATING HORMONE), RHENIUM | | Authors: | Giblin, M.F, Wang, N, Hoffman, T.J, Jurisson, S.S, Quinn, T.P. | | Deposit date: | 1998-11-12 | | Release date: | 1998-11-18 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of alpha-melanotropin peptide analogs cyclized through rhenium and technetium metal coordination.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6DW5
 
 | | SAMHD1 Bound to Gemcitabine-TP in the Catalytic Pocket | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-deoxy-2',2'-difluorocytidine 5'-(tetrahydrogen triphosphate), Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Knecht, K.M, Buzovetsky, O, Schneider, C, Thomas, D, Srikanth, V, Kaderali, L, Tofoleanu, F, Reiss, K, Ferreiros, N, Geisslinger, G, Batista, V.S, Ji, X, Cinatl, J, Keppler, O.T, Xiong, Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6TWG
 
 | | Solution structure of antimicrobial peptide, crabrolin Plus in the presence of Lipopolysaccharide | | Descriptor: | Crabrolin Plus, mutant of Crabrolin peptide | | Authors: | Cantini, F, Bouchemal, N, Savarin, P, Sette, M. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Effect of positive charges in the structural interaction of crabrolin isoforms with lipopolysaccharide.
J.Pept.Sci., 26, 2020
|
|
6UF2
 
 | |
1RRX
 
 | | Crystallographic Evidence for Isomeric Chromophores in 3-Fluorotyrosyl-Green Fluorescent Protein | | Descriptor: | SIGF1-GFP fusion protein | | Authors: | Bae, J.H, Paramita Pal, P, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-12-09 | | Release date: | 2004-06-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Evidence for Isomeric Chromophores in 3-Fluorotyrosyl-Green Fluorescent Protein.
Chembiochem, 5, 2004
|
|
6DQ3
 
 | | Streptococcus pyogenes deacetylase PplD in complex with acetate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IMIDAZOLE, ... | | Authors: | Li, J, Korotkova, N, Korotkov, K.V. | | Deposit date: | 2018-06-10 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | PplD is a de-N-acetylase of the cell wall linkage unit of streptococcal rhamnopolysaccharides
Nat Commun, 13, 2022
|
|
6DQS
 
 | | Class 3 IP3-bound human type 3 1,4,5-inositol trisphosphate receptor | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | Authors: | Hite, R.K, Paknejad, N. | | Deposit date: | 2018-06-11 | | Release date: | 2018-08-01 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DUV
 
 | | Crystal structure of the second bromodomain of human BRD4 in complex with MS417 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, methyl [(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetate | | Authors: | Babault, N, Zhou, M.M. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of Bound Water Molecules in Differential Recognition of Di-Acetylated Histone Peptides by the BRD4 Bromodomains
To be published
|
|
5EU2
 
 | | Crystal structure of murine neuroglobin mutant V101F at ambient pressure | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Colloc'h, N, Girard, E, Vallone, B, Prange, T. | | Deposit date: | 2015-11-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determinants of neuroglobin plasticity highlighted by joint coarse-grained simulations and high pressure crystallography.
Sci Rep, 7, 2017
|
|
