7TQW
 
 | | Kod RSGA incorporating PMT, n+2 | | Descriptor: | DNA polymerase, Primer, Template | | Authors: | Hajjar, M, Chim, N, Chaput, J.C. | | Deposit date: | 2022-01-27 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystallographic analysis of engineered polymerases synthesizing phosphonomethylthreosyl nucleic acid.
Nucleic Acids Res., 50, 2022
|
|
6MU4
 
 | | Bst DNA polymerase I FANA/DNA binary complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(P*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3'), DNA polymerase I, ... | | Authors: | Jackson, L.N, Chim, N, Chaput, J.C. | | Deposit date: | 2018-10-22 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structures of a natural DNA polymerase that functions as an XNA reverse transcriptase.
Nucleic Acids Res., 47, 2019
|
|
6MU5
 
 | | Bst DNA polymerase I TNA/DNA binary complex | | Descriptor: | DNA (5'-D(P*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3'), DNA polymerase I, SULFATE ION, ... | | Authors: | Jackson, L.N, Chim, N, Chaput, J.C. | | Deposit date: | 2018-10-22 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | Crystal structures of a natural DNA polymerase that functions as an XNA reverse transcriptase.
Nucleic Acids Res., 47, 2019
|
|
3LTB
 
 | | X-ray structure of a non-biological ATP binding protein determined in the presence of 10 mM ATP at 2.6 A after 3 weeks of incubation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP BINDING PROTEIN-DX, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Magee, C.L, Allen, J.P, Chaput, J.C. | | Deposit date: | 2010-02-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structures reveal multiple ADP/ATP binding modes for a synthetic class of artificial proteins.
Biochemistry, 49, 2010
|
|
7RSR
 
 | | Kod-RI incorporating PMT, n+2 | | Descriptor: | DNA polymerase, Primer, Template | | Authors: | Hajjar, M, Chim, N, Chaput, J.C. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystallographic analysis of engineered polymerases synthesizing phosphonomethylthreosyl nucleic acid.
Nucleic Acids Res., 50, 2022
|
|
7RSS
 
 | | Kod-RI incorporating DNA, n+2 | | Descriptor: | DNA polymerase, Primer, Template | | Authors: | Hajjar, M, Chim, N, Chaput, J.C. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystallographic analysis of engineered polymerases synthesizing phosphonomethylthreosyl nucleic acid.
Nucleic Acids Res., 50, 2022
|
|
6DSV
 
 | | Bst DNA polymerase I post-chemistry (n+2) structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
6DSW
 
 | | Bst DNA polymerase I pre-chemistry (n) structure | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*G)-3'), DNA (5'-D(P*AP*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
6DSY
 
 | | Bst DNA polymerase I post-chemistry (n+1) structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3'), DNA (5'-D(P*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
6DSU
 
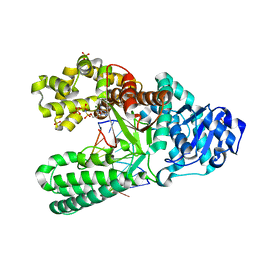 | | Bst DNA polymerase I pre-insertion complex structure | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
6DSX
 
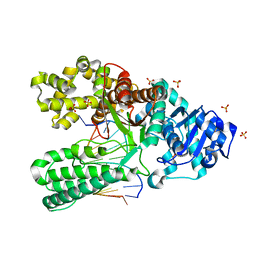 | | Bst DNA polymerase I post-chemistry (n+1 with dATP soak) structure | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3'), DNA polymerase I, ... | | Authors: | Chim, N, Jackson, L.N, Chaput, J.C. | | Deposit date: | 2018-06-14 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structures of DNA polymerase I capture novel intermediates in the DNA synthesis pathway.
Elife, 7, 2018
|
|
2LUJ
 
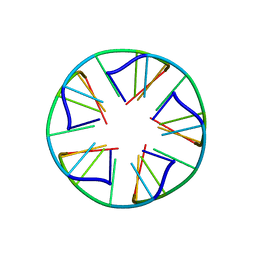 | | Solution structure of a parallel-stranded oligoisoguanine DNA pentaplex formed by d(T(iG)4T) in the presence of Cs ions | | Descriptor: | DNA (5'-D(*TP*(IGU)P*(IGU)P*(IGU)P*(IGU)P*T)-3') | | Authors: | Kang, M, Heuberger, B, Chaput, J.C, Switzer, C, Feigon, J. | | Deposit date: | 2012-06-14 | | Release date: | 2012-07-25 | | Last modified: | 2016-11-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Parallel-Stranded Oligoisoguanine DNA Pentaplex Formed by d(T(iG)4T) in the Presence of Cs(+) Ions.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
2P0X
 
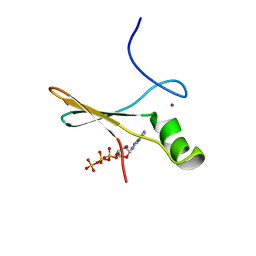 | | solution structure of a non-biological ATP-binding protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ZINC ION, abiotic ATP-binding, ... | | Authors: | Mansy, S.S, Szostak, J.W, Chaput, J.C. | | Deposit date: | 2007-03-01 | | Release date: | 2007-08-07 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure and Evolutionary Analysis of a Non-biological ATP-binding Protein
J.Mol.Biol., 371, 2007
|
|
2P09
 
 | | Structural Insights into the Evolution of a Non-Biological Protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Smith, M, Rosenow, M, Wang, M, Allen, J.P, Szostak, J.W, Chaput, J.C. | | Deposit date: | 2007-02-28 | | Release date: | 2007-06-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into the evolution of a non-biological protein: importance of surface residues in protein fold optimization.
PLoS ONE, 2, 2007
|
|
2P05
 
 | | Structural Insights into the Evolution of a Non-Biological Protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Smith, M, Rosenow, M, Wang, M, Allen, J.P, Szostak, J.W, Chaput, J.C. | | Deposit date: | 2007-02-28 | | Release date: | 2007-06-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the evolution of a non-biological protein: importance of surface residues in protein fold optimization.
PLoS ONE, 2, 2007
|
|
3DGO
 
 | | A non-biological ATP binding protein with a Tyr-Phe mutation in the ligand binding domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP Binding Protein-DX, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Allen, J.P, Chaput, J.C. | | Deposit date: | 2008-06-13 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A synthetic protein selected for ligand binding affinity mediates ATP hydrolysis.
Acs Chem.Biol., 4, 2009
|
|
3DGM
 
 | |
3DGN
 
 | | A non-biological ATP binding protein crystallized in the presence of 100 mM ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP Binding Protein-DX, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Simmons, C.R, Allen, J.P, Chaput, J.C. | | Deposit date: | 2008-06-13 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A synthetic protein selected for ligand binding affinity mediates ATP hydrolysis.
Acs Chem.Biol., 4, 2009
|
|
3DGL
 
 | | 1.8 A Crystal Structure of a Non-biological Protein with Bound ATP in a Novel Bent Conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP Binding Protein-DX, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Simmons, C.R, Allen, J.P, Chaput, J.C. | | Deposit date: | 2008-06-13 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A synthetic protein selected for ligand binding affinity mediates ATP hydrolysis.
Acs Chem.Biol., 4, 2009
|
|
5VU5
 
 | | TNA polymerase, apo | | Descriptor: | DNA polymerase | | Authors: | Chim, N, Chaput, J.C. | | Deposit date: | 2017-05-18 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for TNA synthesis by an engineered TNA polymerase.
Nat Commun, 8, 2017
|
|
5VU7
 
 | | TNA polymerase, open ternary complex | | Descriptor: | 5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXYTETRAHYDROFURAN-3-YL DIHYDROGEN PHOSPHATE, DNA polymerase, DNA template, ... | | Authors: | Chim, N, Chaput, J.C. | | Deposit date: | 2017-05-18 | | Release date: | 2017-12-06 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis for TNA synthesis by an engineered TNA polymerase.
Nat Commun, 8, 2017
|
|
5VU9
 
 | | TNA polymerase, translocated product | | Descriptor: | DNA polymerase, DNA template, DNA/TNA hybrid primer | | Authors: | Chim, N, Chaput, J.C. | | Deposit date: | 2017-05-18 | | Release date: | 2017-12-06 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for TNA synthesis by an engineered TNA polymerase.
Nat Commun, 8, 2017
|
|
5VU8
 
 | | TNA polymerase, closed ternary complex | | Descriptor: | DNA polymerase, DNA template, DNA/TNA hybrid primer, ... | | Authors: | Chim, N, Chaput, J.C. | | Deposit date: | 2017-05-18 | | Release date: | 2017-12-06 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for TNA synthesis by an engineered TNA polymerase.
Nat Commun, 8, 2017
|
|
5VU6
 
 | |
3LTD
 
 | | X-ray structure of a non-biological ATP binding protein determined at 2.8 A by multi-wavelength anomalous dispersion | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP BINDING PROTEIN-DX, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Magee, C.L, Allen, J.P, Chaput, J.C. | | Deposit date: | 2010-02-15 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structures reveal multiple ADP/ATP binding modes for a synthetic class of artificial proteins.
Biochemistry, 49, 2010
|
|
