3OIL
 
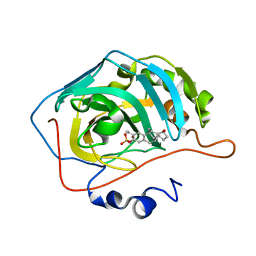 | | Human Carbonic anhydrase II mutant A65S, N67Q (CA IX mimic) bound by 2-Ethylestradiol 3-O-sulfamate | | Descriptor: | (14beta,17alpha)-2-ethyl-17-hydroxyestra-1(10),2,4-trien-3-yl sulfamate, Carbonic anhydrase 2, ZINC ION | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
3NWM
 
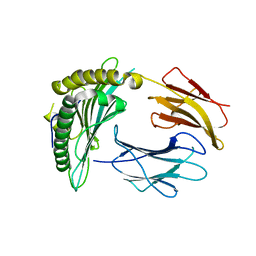 | | Crystal structure of a single chain construct composed of MHC class I H-2Kd, beta-2microglobulin and a peptide which is an autoantigen for type 1 diabetes | | Descriptor: | Peptide/beta-2microglobulin/MHC class I H-2Kd chimeric protein | | Authors: | Ramagopal, U.A, Samanta, D, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-07-09 | | Release date: | 2011-07-06 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterization of a single-chain peptide-MHC molecule that modulates both naive and activated CD8+ T cells.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3O3I
 
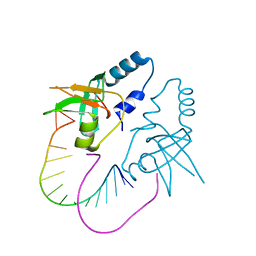 | | Crystal Structure of human Hiwi1 PAZ domain (residues 277-399) in complex with 14-mer RNA (12-bp + 2-nt overhang) containing 2'-OH at its 3'-end | | Descriptor: | Piwi-like protein 1, RNA (5'-R(*GP*CP*GP*AP*AP*UP*AP*UP*UP*CP*GP*CP*UP*U)-3') | | Authors: | Tian, Y, Simanshu, D.K, Ma, J.-B, Patel, D.J. | | Deposit date: | 2010-07-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OKV
 
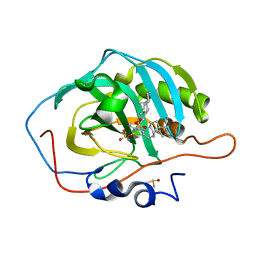 | | Human Carbonic Anhydrase II A65S, N67Q (CA IX mimic) bound with 2-Ethylestrone 3-O-sulfamate | | Descriptor: | (9beta)-2-ethyl-17-oxoestra-1(10),2,4-trien-3-yl sulfamate, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-25 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
3OIM
 
 | | Human Carbonic anhydrase II bound by 2-Ethylestradiol 3-O-sulfamate | | Descriptor: | (14beta,17alpha)-2-ethyl-17-hydroxyestra-1(10),2,4-trien-3-yl sulfamate, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
3OKU
 
 | | Human Carbonic Anhydrase II in complex with 2-Ethylestrone-3-O-sulfamate | | Descriptor: | (9beta)-2-ethyl-17-oxoestra-1(10),2,4-trien-3-yl sulfamate, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-25 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
2KY5
 
 | | Solution structure of the PECAM-1 cytoplasmic tail with DPC | | Descriptor: | Platelet endothelial cell adhesion molecule | | Authors: | Lytle, B.L, Peterson, F.C, Volkman, B.F, Paddock, C, Newman, D.K, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-05-14 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Residues within a lipid-associated segment of the PECAM-1 cytoplasmic domain are susceptible to inducible, sequential phosphorylation.
Blood, 117, 2011
|
|
2HTA
 
 | | Crystal Structure of a putative mutarotase (YeaD) from Salmonella typhimurium in orthorhombic form | | Descriptor: | GLYCEROL, Putative enzyme related to aldose 1-epimerase, SULFATE ION | | Authors: | Chittori, S, Simanshu, D.K, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2006-07-25 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the putative mutarotase YeaD from Salmonella typhimurium: structural comparison with galactose mutarotases.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3O7V
 
 | | Crystal Structure of human Hiwi1 (V361M) PAZ domain (residues 277-399) in complex with 14-mer RNA (12-bp + 2-nt overhang) containing 2'-OCH3 at its 3'-end | | Descriptor: | Piwi-like protein 1, RNA (5'-R(*GP*CP*GP*AP*AP*UP*AP*UP*UP*CP*GP*CP*UP*(OMU))-3') | | Authors: | Tian, Y, Simanshu, D.K, Ma, J.-B, Patel, D.J. | | Deposit date: | 2010-08-01 | | Release date: | 2011-01-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3O6E
 
 | | Crystal Structure of human Hiwi1 PAZ domain (residues 277-399) in complex with 14-mer RNA (12-bp + 2-nt overhang) containing 2'-OCH3 at its 3'-end | | Descriptor: | Piwi-like protein 1, RNA (5'-R(*GP*CP*GP*AP*AP*UP*AP*UP*UP*CP*GP*CP*UP*(OMU))-3') | | Authors: | Tian, Y, Simanshu, D.K, Ma, J.-B, Patel, D.J. | | Deposit date: | 2010-07-28 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Inaugural Article: Structural basis for piRNA 2'-O-methylated 3'-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2JO5
 
 | | Tetrameric structure of KIA7F peptide | | Descriptor: | KIA7F | | Authors: | Lopez de la Osa, J, Gonzalez, C, Laurents, D.V, Chakrabartty, A, Bateman, D.A. | | Deposit date: | 2007-02-21 | | Release date: | 2007-09-18 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Getting specificity from simplicity in putative proteins from the prebiotic earth.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3SRZ
 
 | | Clostridium difficile toxin A (TcdA) glucolsyltransferase domain bound to UDP-glucose | | Descriptor: | MANGANESE (II) ION, Toxin A, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Pruitt, R.N, Chumbler, N.M, Farrow, M.A, Seeback, S.A, Friedman, D.B, Spiller, B.W, Lacy, D.B. | | Deposit date: | 2011-07-07 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural Determinants of Clostridium difficile Toxin A Glucosyltransferase Activity.
J.Biol.Chem., 287, 2012
|
|
6OB3
 
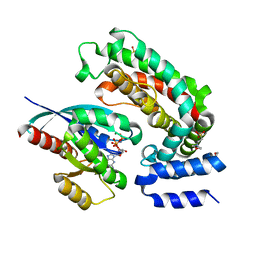 | | Crystal structure of G13D-KRAS (GMPPNP-bound) in complex with GAP-related domain (GRD) of neurofibromin (NF1) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GTPase KRas, ... | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2019-03-19 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | KRAS G13D sensitivity to neurofibromin-mediated GTP hydrolysis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5A3F
 
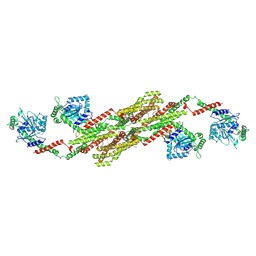 | | Crystal structure of the dynamin tetramer | | Descriptor: | DYNAMIN 3 | | Authors: | Reubold, T.F, Faelber, K, Plattner, N, Posor, Y, Branz, K, Curth, U, Schlegel, J, Anand, R, Manstein, D.J, Noe, F, Haucke, V, Daumke, O, Eschenburg, S. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of the Dynamin Tetramer
Nature, 525, 2015
|
|
5AAZ
 
 | | TBK1 recruitment to cytosol-invading Salmonella induces anti- bacterial autophagy | | Descriptor: | OPTINEURIN, ZINC ION | | Authors: | Thurston, T.l, Allen, M.D, Ravenhill, B, Karpiyevitch, M, Bloor, S, Kaul, A, Matthews, S, Komander, D, Holden, D, Bycroft, M, Randow, F. | | Deposit date: | 2015-07-31 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Recruitment of Tbk1 to Cytosol-Invading Salmonella Induces Wipi2-Dependent Antibacterial Autophagy.
Embo J., 35, 2016
|
|
1AS2
 
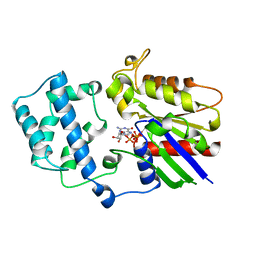 | | GDP+PI BOUND G42V GIA1 | | Descriptor: | GIA1, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION | | Authors: | Raw, A.S, Coleman, D.E, Gilman, A.G, Sprang, S.R. | | Deposit date: | 1997-08-11 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical characterization of the GTPgammaS-, GDP.Pi-, and GDP-bound forms of a GTPase-deficient Gly42 --> Val mutant of Gialpha1.
Biochemistry, 36, 1997
|
|
2BVS
 
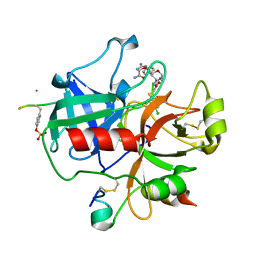 | | Human thrombin complexed with fragment-based small molecules occupying the S1 pocket | | Descriptor: | ALPHA THROMBIN, HIRUDIN VARIANT-2, N-[2-(2-CARBAMOYLMETHOXY-ETHOXY)-ETHYL]-2-[2-(4-CHLORO-PHENYLSULFANYL)-ACETYLAMINO]-3-(4-GUANIDINO-PHENYL)-PROPIONAMIDE | | Authors: | Neumann, T, Junker, H.-D, Keil, O, Burkert, K, Ottleben, H, Gamer, J, Sekul, R, Deppe, H, Feurer, A, Tomandl, D, Metz, G. | | Deposit date: | 2005-07-04 | | Release date: | 2006-10-25 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of Thrombin Inhibitor Fragments from Chemical Microarray Screening
Lett.Drug Des.Discovery, 2, 2005
|
|
1M3A
 
 | | Solution structure of a circular form of the truncated N-terminal SH3 domain from oncogene protein c-Crk. | | Descriptor: | Proto-oncogene C-crk | | Authors: | Schumann, F.H, Varadan, R, Tayakuniyil, P.P, Hall, J.B, Camarero, J.A, Fushman, D. | | Deposit date: | 2002-06-27 | | Release date: | 2003-08-05 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Changing protein backbone topology: Structural and dynamic consequences of the backbone cyclization in SH3 domain
To be Published
|
|
2EU3
 
 | | Human Carbonic anhydrase II in complex with novel inhibitors | | Descriptor: | 1-(5-AMINO-1,3,4-THIADIAZOL-2-YL)-1,1-DIFLUOROMETHANESULFONAMIDE, Carbonic anhydrase 2, ZINC ION | | Authors: | Fisher, S.Z, Govindasamy, L, Boyle, N, Agbandje-McKenna, M, Silverman, D.N, Blackburn, G.M, McKenna, R. | | Deposit date: | 2005-10-28 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies reveal that the incorporation of spacer groups in carbonic anhydrase inhibitors causes alternate binding modes.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
3GYU
 
 | | Nuclear receptor DAF-12 from parasitic nematode Strongyloides stercoralis in complex with its physiological ligand dafachronic acid delta 7 | | Descriptor: | (5beta,14beta,17alpha,25R)-3-oxocholest-7-en-26-oic acid, Nuclear hormone receptor of the steroid/thyroid hormone receptors superfamily, SRC1 | | Authors: | Zhou, X.E, Wang, Z, Suino-Powell, K, Motola, D.L, Conneely, A, Ogata, C, Sharma, K.K, Auchus, R.J, Kliewer, S.A, Xu, H.E, Mangelsdorf, D.J. | | Deposit date: | 2009-04-05 | | Release date: | 2009-07-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the nuclear receptor DAF-12 as a therapeutic target in parasitic nematodes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4KNC
 
 | | Structural and functional characterization of Pseudomonas aeruginosa AlgX | | Descriptor: | Alginate biosynthesis protein AlgX | | Authors: | Riley, L.M, Weadge, J.T, Baker, P, Robinson, H, Codee, J.D.C, Tipton, P.A, Ohman, D.E, Howell, P.L. | | Deposit date: | 2013-05-09 | | Release date: | 2013-06-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural and Functional Characterization of Pseudomonas aeruginosa AlgX: ROLE OF AlgX IN ALGINATE ACETYLATION.
J.Biol.Chem., 288, 2013
|
|
6OB2
 
 | | Crystal structure of wild-type KRAS (GMPPNP-bound) in complex with GAP-related domain (GRD) of neurofibromin (NF1) | | Descriptor: | CHLORIDE ION, GLYCEROL, GTPase KRas, ... | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2019-03-19 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.845 Å) | | Cite: | KRAS G13D sensitivity to neurofibromin-mediated GTP hydrolysis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2BVR
 
 | | Human thrombin complexed with fragment-based small molecules occupying the S1 pocket | | Descriptor: | 2-[2-(4-CHLORO-PHENYLSULFANYL)-ACETYLAMINO]-3-(4-GUANIDINO-PHENYL)-PROPIONAMIDE, ALPHA THROMBIN, HIRUDIN VARIANT-2 | | Authors: | Neumann, T, Junker, H.-D, Keil, O, Burkert, K, Ottleben, H, Gamer, J, Sekul, R, Deppe, H, Feurer, A, Tomandl, D, Metz, G. | | Deposit date: | 2005-07-04 | | Release date: | 2006-10-25 | | Last modified: | 2016-12-21 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of Thrombin Inhibitor Fragments from Chemical Microarray Screening
Lett.Drug Des.Discovery, 2, 2005
|
|
7N62
 
 | | SARS-CoV-2 Spike (2P) in complex with C12C9 Fab (NTD local reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C12C9 Fab heavy chain, C12C9 Fab light chain, ... | | Authors: | Windsor, I.W, Jenni, S, Bajic, G, Tong, P, Gautam, A.K, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-06-07 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike.
Biorxiv, 2021
|
|
4KVR
 
 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase (mutant V41Y) | | Descriptor: | Aldehyde decarbonylase, FE (III) ION, HEXANOIC ACID | | Authors: | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
