8E0S
 
 | | DAHP (3-deoxy-D-arabinoheptulosonate-7-phosphate) Synthase complexed with DAHP Oxime in unbound:(bound)2:unbound conformations | | Descriptor: | DAHP Oxime, Phospho-2-dehydro-3-deoxyheptonate aldolase, Phe-sensitive, ... | | Authors: | Berti, P.J, Junop, M.S, Grainger, R. | | Deposit date: | 2022-08-09 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Role of Half-of-Sites Reactivity and Inter-Subunit Communications in DAHP Synthase Catalysis and Regulation.
Biochemistry, 61, 2022
|
|
2R69
 
 | | Crystal structure of Fab 1A1D-2 complexed with E-DIII of Dengue virus at 3.8 angstrom resolution | | Descriptor: | Heavy chain of 1A1D-2, Light chain of 1A1D-2, Major envelope protein E | | Authors: | Lok, S.M, Kostyuchenko, V.K, Nybakken, G.E, Holdaway, H.A, Battisti, A.J, Sukupolvi-petty, S, Sedlak, D, Fremont, D.H, Chipman, P.R, Roehrig, J.T, Diamond, M.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-09-05 | | Release date: | 2007-12-25 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins.
Nat.Struct.Mol.Biol., 15, 2008
|
|
6A3F
 
 | | Levoglucosan dehydrogenase, apo form | | Descriptor: | Putative dehydrogenase, SULFATE ION | | Authors: | Sugiura, M, Yamada, C, Arakawa, T, Fushinobu, S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification, functional characterization, and crystal structure determination of bacterial levoglucosan dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6NCH
 
 | | Crystal structure of CDP-Chase: Raster data collection | | Descriptor: | D-ribose, PHOSPHATE ION, Phosphohydrolase (MutT/nudix family protein), ... | | Authors: | Miller, M.S, Shi, W, Gabelli, S.B. | | Deposit date: | 2018-12-11 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
5L39
 
 | |
2R5B
 
 | | Structure of the gp41 N-trimer in complex with the HIV entry inhibitor PIE7 | | Descriptor: | HIV entry inhibitor PIE7, SULFATE ION, gp41 N-peptide | | Authors: | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | Deposit date: | 2007-09-03 | | Release date: | 2007-10-02 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R5D
 
 | | Structure of the gp41 N-trimer in complex with the HIV entry inhibitor PIE7 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, HIV entry inhibitor PIE7, ... | | Authors: | VanDemark, A.P, Welch, B, Heroux, A, Hill, C.P, Kay, M.S. | | Deposit date: | 2007-09-03 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Potent D-peptide inhibitors of HIV-1 entry
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R5O
 
 | | Crystal structure of the C-terminal domain of wzt | | Descriptor: | CHLORIDE ION, Putative ATP binding component of ABC-transporter, SODIUM ION, ... | | Authors: | Kimber, M.S, Cuthbertson, L, Whitfield, C. | | Deposit date: | 2007-09-04 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Substrate binding by a bacterial ABC transporter involved in polysaccharide export.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R8P
 
 | | Transketolase from E. coli in complex with substrate D-fructose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-C-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium-2-yl}-6-O-phosphono-D-glucitol, CALCIUM ION, ... | | Authors: | Wille, G, Asztalos, P, Weiss, M.S, Tittmann, K. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Strain and near attack conformers in enzymic thiamin catalysis: X-ray crystallographic snapshots of bacterial transketolase in covalent complex with donor ketoses xylulose 5-phosphate and fructose 6-phosphate, and in noncovalent complex with acceptor aldose ribose 5-phosphate.
Biochemistry, 46, 2007
|
|
6A1C
 
 | | Crystal structure of the CK2a1-go289 complex | | Descriptor: | 1,2-ETHANEDIOL, 5-bromanyl-2-methoxy-4-[(E)-(3-methylsulfanyl-5-phenyl-1,2,4-triazol-4-yl)iminomethyl]phenol, Casein kinase II subunit alpha, ... | | Authors: | Kinoshita, T, Tsuyuguchi, M. | | Deposit date: | 2018-06-07 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Cell-based screen identifies a new potent and highly selective CK2 inhibitor for modulation of circadian rhythms and cancer cell growth.
Sci Adv, 5, 2019
|
|
6A22
 
 | | Ternary complex of Human ROR gamma Ligand Binding Domain With Compound T. | | Descriptor: | 2-[2-[1-~{tert}-butyl-5-(4-methoxyphenyl)pyrazol-4-yl]-1,3-thiazol-4-yl]-~{N}-(oxan-4-ylmethyl)ethanamide, Nuclear receptor ROR-gamma, Nuclear receptor corepressor 2 | | Authors: | Noguchi, M, Nomura, A, Doi, S, Yamaguchi, K, Adachi, T. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Ternary crystal structure of human ROR gamma ligand-binding-domain, an inhibitor and corepressor peptide provides a new insight into corepressor interaction
Sci Rep, 8, 2018
|
|
6A3I
 
 | | Levoglucosan dehydrogenase, complex with NADH and levoglucosan | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Levoglucosan, Putative dehydrogenase | | Authors: | Sugiura, M, Yamada, C, Arakawa, T, Fushinobu, S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Identification, functional characterization, and crystal structure determination of bacterial levoglucosan dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
2R50
 
 | |
3C27
 
 | | Cyanofluorophenylacetamides as Orally Efficacious Thrombin Inhibitors | | Descriptor: | Hirudin-3A, N-[2-(carbamimidamidooxy)ethyl]-2-{6-cyano-3-[(2,2-difluoro-2-pyridin-2-ylethyl)amino]-2-fluorophenyl}acetamide, Thrombin heavy chain, ... | | Authors: | Spurlino, J.C, McMillan, M, Lewandowski, F, Milligan, C. | | Deposit date: | 2008-01-24 | | Release date: | 2009-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Cyanofluorophenylacetamides as Orally Efficacious Thrombin Inhibitors
To be Published
|
|
6JNO
 
 | | RXRa structure complexed with CU-6PMN | | Descriptor: | 7-oxidanyl-2-oxidanylidene-6-(3,5,5,8,8-pentamethyl-6,7-dihydronaphthalen-2-yl)chromene-3-carboxylic acid, Retinoic acid receptor RXR-alpha | | Authors: | Kawasaki, M, Nakano, S, Motoyama, T, Yamada, S, Watanabe, M, Takamura, Y, Fujihara, M, Tokiwa, H, Kakuta, H, Ito, S. | | Deposit date: | 2019-03-17 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Competitive Binding Assay with an Umbelliferone-Based Fluorescent Rexinoid for Retinoid X Receptor Ligand Screening.
J.Med.Chem., 62, 2019
|
|
8R6W
 
 | | Structure of the SFTSV L protein in a transcription-priming state with bound capped RNA [TRANSCRIPTION-PRIMING] | | Descriptor: | RNA (5'-R(*(M7G)*AP*AP*A)-3'), RNA (5'-R(*AP*CP*AP*C)-3'), RNA (5'-R(*AP*CP*AP*CP*AP*GP*AP*GP*AP*CP*GP*CP*CP*CP*AP*G)-3'), ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Busch, C, Milewski, M, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2023-11-23 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural snapshots of phenuivirus cap-snatching and transcription.
Nucleic Acids Res., 52, 2024
|
|
8R6Y
 
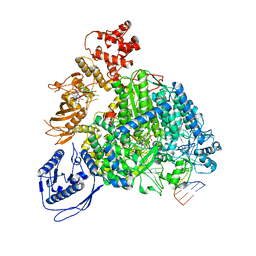 | | Structure of the SFTSV L protein stalled in a transcription-specific early elongation state with bound capped RNA [TRANSCRIPTION-EARLY-ELONGATION] | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, RNA (5'-R(*(M7G)*AP*AP*A)-3'), ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Busch, C, Milewski, M, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2023-11-23 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural snapshots of phenuivirus cap-snatching and transcription.
Nucleic Acids Res., 52, 2024
|
|
8IHO
 
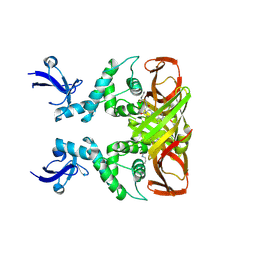 | | Crystal structures of SARS-CoV-2 papain-like protease in complex with covalent inhibitors | | Descriptor: | Papain-like protease nsp3, ZINC ION, covalent inhibitor | | Authors: | Wang, Q, Hu, H, Li, M, Xu, Y. | | Deposit date: | 2023-02-23 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-Based Design of Potent Peptidomimetic Inhibitors Covalently Targeting SARS-CoV-2 Papain-like Protease.
Int J Mol Sci, 24, 2023
|
|
8R6U
 
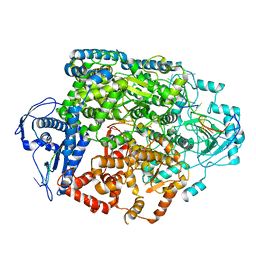 | | Structure of the SFTSV L protein in a transcription-priming state without capped RNA [TRANSCRIPTION-PRIMING (in vitro)] | | Descriptor: | MAGNESIUM ION, RNA (5'-R(P*CP*UP*GP*GP*GP*CP*GP*GP*UP*CP*UP*UP*UP*GP*UP*GP*U)-3'), RNA primer, ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Busch, C, Milewski, M, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2023-11-23 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural snapshots of phenuivirus cap-snatching and transcription.
Nucleic Acids Res., 52, 2024
|
|
2VJY
 
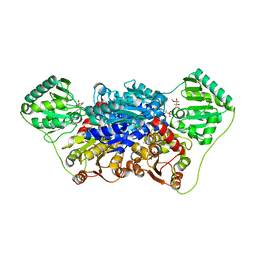 | | Pyruvate decarboxylase from Kluyveromyces lactis in complex with the substrate analogue methyl acetylphosphonate | | Descriptor: | MAGNESIUM ION, METHYL HYDROGEN (S)-ACETYLPHOSPHONATE, PYRUVATE DECARBOXYLASE, ... | | Authors: | Kutter, S, Wille, G, Weiss, M.S, Konig, S. | | Deposit date: | 2007-12-14 | | Release date: | 2009-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Covalently Bound Substrate at the Regulatory Site of Yeast Pyruvate Decarboxylases Triggers Allosteric Enzyme Activation.
J.Biol.Chem., 284, 2009
|
|
8R41
 
 | | Structure of CHI3L1 in complex with inhibitor 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | Authors: | Nowak, E, Napiorkowska-Gromadzka, A, Nowotny, M. | | Deposit date: | 2023-11-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Discovery of High-Affinity Small Molecule Ligands and Development of Tool Probes to Study the Role of Chitinase-3-Like Protein 1.
J.Med.Chem., 67, 2024
|
|
8HYF
 
 | | Crystal Structure of Banana Lectin In-complex with Fucose at 2.95 A Resolution | | Descriptor: | CADMIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Rasheed, S, Arif, R, Huda, N, Ahmad, M.S, Mateen, S.M. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of Banana Lectin in-Complex with L-Fucose at 2.95 A Resolution
To Be Published
|
|
8I3E
 
 | |
6ND6
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with erythromycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.85A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Plessa, E, Chen, C.-W, Bougas, A, Krokidis, M.G, Dinos, G.P, Polikanov, Y.S. | | Deposit date: | 2018-12-13 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition.
RNA, 25, 2019
|
|
2VF3
 
 | | Aquifex aeolicus IspE in complex with ligand | | Descriptor: | 4-DIPHOSPHOCYTIDYL-2C-METHYL-D-ERYTHRITOL KINASE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Hunter, W.N. | | Deposit date: | 2007-10-30 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of the Kinase Ispe: Structure-Activity Relationships and Co-Crystal Structure Analysis.
Org.Biomol.Chem., 6, 2008
|
|
