6D8V
 
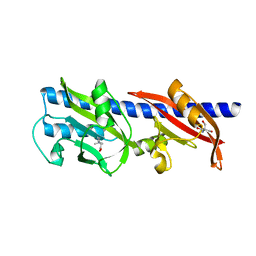 | | Methyl-accepting Chemotaxis protein X | | Descriptor: | 1,1-DIMETHYL-PROLINIUM, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Probable chemoreceptor (Methyl-accepting chemotaxis) transmembrane protein | | Authors: | Shrestha, M, Schubot, F.D. | | Deposit date: | 2018-04-27 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the sensory domain of McpX fromSinorhizobium meliloti, the first known bacterial chemotactic sensor for quaternary ammonium compounds.
Biochem. J., 475, 2018
|
|
4OW3
 
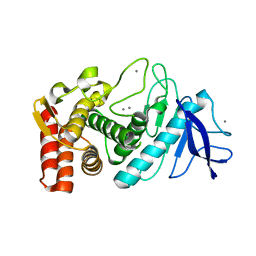 | | Thermolysin structure determined by free-electron laser | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Hattne, J, Echols, N, Tran, R, Kern, J, Gildea, R.J, Brewster, A.S, Alonso-Mori, R, Glockner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, White, W.E, Schafer, D.W, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Glatzel, P, Zwart, P.H, Grosse-Kunstleve, R.W, Bogan, M.J, Messerschmidt, M, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Yano, J, Bergmann, U, Yachandra, V.K, Adams, P.D, Sauter, N.K. | | Deposit date: | 2014-01-30 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Accurate macromolecular structures using minimal measurements from X-ray free-electron lasers.
Nat.Methods, 11, 2014
|
|
8K2C
 
 | | Cryo-EM structure of the human 80S ribosome with Tigecycline | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Li, X, Wang, M, Cheng, J. | | Deposit date: | 2023-07-12 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for differential inhibition of eukaryotic ribosomes by tigecycline.
Nat Commun, 15, 2024
|
|
6D95
 
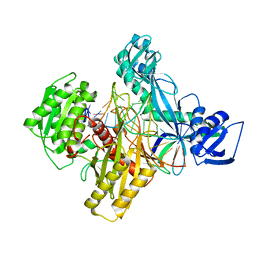 | | Ternary RsAgo Complex with Guide RNA Paired and Target DNA containing A8-A8' Non-Canonical Pair | | Descriptor: | DNA 24-Mer, MAGNESIUM ION, RNA (5'-R(P*UP*UP*AP*CP*UP*GP*CP*AP*CP*AP*GP*GP*UP*GP*AP*CP*GP*A)-3'), ... | | Authors: | Liu, Y, Esyunina, D, Olovnikov, I, Teplova, M, Patel, D.J. | | Deposit date: | 2018-04-27 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Accommodation of Helical Imperfections in Rhodobacter sphaeroides Argonaute Ternary Complexes with Guide RNA and Target DNA.
Cell Rep, 24, 2018
|
|
8K7N
 
 | | Structure of ferric-binding protein KfuA | | Descriptor: | FE (III) ION, Ferric iron ABC transporter, iron-binding protein | | Authors: | Zhao, Q, Bartlam, M. | | Deposit date: | 2023-07-26 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Klebsiella pneumoniae ferric-binding protein KfuA
To be published
|
|
3LK6
 
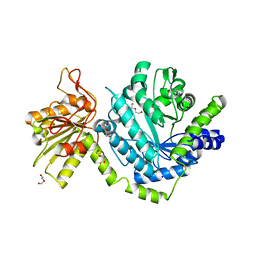 | |
6DB0
 
 | | N-Terminal bromodomain of Human BRD2 with a Tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2S,4R)-4-[(2-chlorophenyl)amino]-2-methyl-6-(1H-pyrazol-3-yl)-3,4-dihydroquinolin-1(2H)-yl]ethan-1-one, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2018-05-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | N-Terminal bromodomain of Human BRD2 with a Tetrahydroquinoline analogue
To Be Published
|
|
6DBC
 
 | | Second bromodomain of Human BRD2 with a Tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2S,4R)-4-[(2-chlorophenyl)amino]-2-methyl-6-(1H-pyrazol-3-yl)-3,4-dihydroquinolin-1(2H)-yl]ethan-1-one, Bromodomain-containing protein 2 | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2018-05-03 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Second bromodomain of Human BRD2 with a Tetrahydroquinoline analogue
To Be Published
|
|
4OC7
 
 | | Retinoic acid receptor alpha in complex with (E)-3-(3'-allyl-6-hydroxy-[1,1'-biphenyl]-3-yl)acrylic acid and a fragment of the coactivator TIF2 | | Descriptor: | (2E)-3-[6-hydroxy-3'-(prop-2-en-1-yl)biphenyl-3-yl]prop-2-enoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Leysen, S, Scheepstra, M, Brunsveld, L, Milroy, L.G, Ottmann, C. | | Deposit date: | 2014-01-08 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A natural-product switch for a dynamic protein interface.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4OCD
 
 | | In and Out the minor groove: Interaction of an AT rich-DNA with the CD27 drug | | Descriptor: | N1-(4,5-dihydro-1H-imidazol-2-yl)-N4-(4-((4,5-dihydro-1H-imidazol-2-yl)amino)phenyl)benzene-1,4-diamine, d(AAAATTTT) | | Authors: | Acosta-Reyes, F.J, Dardonville, C, de Koning, H.P, Natto, M, Subirana, J.A, Campos, J.L. | | Deposit date: | 2014-01-08 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | In and out of the minor groove: interaction of an AT-rich DNA with the drug CD27
Acta Crystallogr.,Sect.D, D70, 2014
|
|
6DBI
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS nicked DNA intermediates | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS signal end, Forward strand of 23-RSS signal end, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBU
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3L6X
 
 | | Crystal structure of p120 catenin in complex with E-cadherin | | Descriptor: | Catenin delta-1, E-cadherin, SULFATE ION | | Authors: | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | Deposit date: | 2009-12-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
6DD3
 
 | | Crystal structure of the double mutant (D52N/D407A) of NT5C2-537X in the active state | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-09 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DE3
 
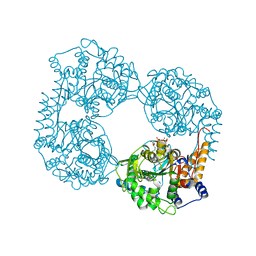 | | Crystal structure of the double mutant (R39Q/D52N) of the full-length NT5C2 in the active state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytosolic purine 5'-nucleotidase, MAGNESIUM ION, ... | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L. | | Deposit date: | 2018-05-11 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
6DDC
 
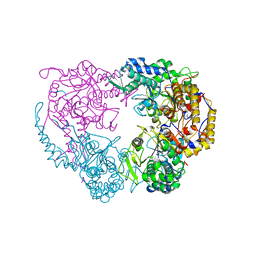 | | Crystal structure of the single mutant (D52N) of NT5C2-537X in the basal state, Northeast Structural Genomics Consortium Target | | Descriptor: | Cytosolic purine 5'-nucleotidase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Forouhar, F, Dieck, C.L, Tzoneva, G, Carpenter, Z, Ambesi-Impiombato, A, Sanchez-Martin, M, Kirschner-Schwabe, R, Lew, S, Seetharaman, J, Ferrando, A.A, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-05-09 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure and Mechanisms of NT5C2 Mutations Driving Thiopurine Resistance in Relapsed Lymphoblastic Leukemia.
Cancer Cell, 34, 2018
|
|
8K96
 
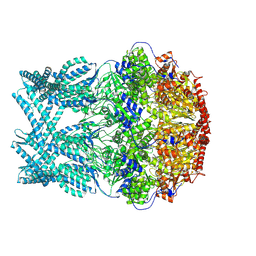 | | CryoEM structure of LonC protease hepatmer with Bortezomib | | Descriptor: | N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, PHOSPHATE ION, endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
6DEB
 
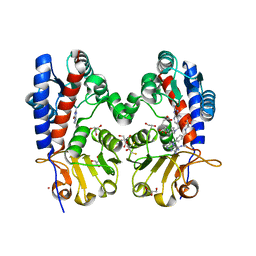 | | Crystal Structure of Bifunctional Enzyme FolD-Methylenetetrahydrofolate Dehydrogenase/Cyclohydrolase in the Complex with Methotrexate from Campylobacter jejuni | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional protein FolD, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Maltseva, N, Grimshaw, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-11 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Bifunctional Enzyme FolD-Methylenetetrahydrofolate Dehydrogenase/Cyclohydrolase in the Complex with Methotrexate from Campylobacter jejuni
To Be Published
|
|
4OGP
 
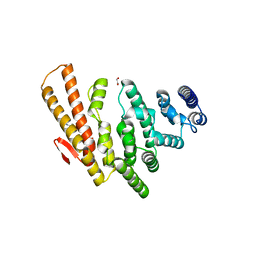 | | Structure of C-terminal domain from S. cerevisiae Pat1 decapping activator (Space group : P21) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA topoisomerase 2-associated protein PAT1 | | Authors: | Fourati-Kammoun, Z, Kolesnikova, O, Back, R, Keller, J, Lazar, N, Gaudon-Plesse, C, Seraphin, B, Graille, M. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The C-terminal domain from S. cerevisiae Pat1 displays two conserved regions involved in decapping factor recruitment.
Plos One, 9, 2014
|
|
8KAQ
 
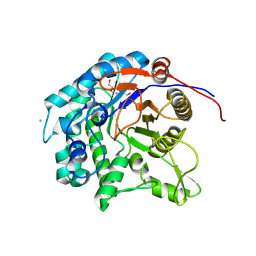 | | Glycoside hydrolase family 1 beta-glucosidase (E318G mutant) from Streptomyces griseus (sophorose complex) | | Descriptor: | CHLORIDE ION, Putative beta-glucosidase, TRIETHYLENE GLYCOL, ... | | Authors: | Kumakura, H, Motouchi, S, Nakai, H, Nakajima, M. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Identification of beta-1,2-glucan-associated glycoside hydrolase family 1 beta-glucosidase from Streptomyces griseus
To Be Published
|
|
4OH0
 
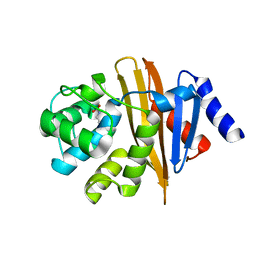 | | Crystal structure of OXA-58 carbapenemase | | Descriptor: | Beta-lactamase OXA-58, CHLORIDE ION | | Authors: | Smith, C.A, Antunes, N.T, Toth, M, Vakulenko, S.B. | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-26 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Carbapenemase OXA-58 from Acinetobacter baumannii.
Antimicrob.Agents Chemother., 58, 2014
|
|
3LIX
 
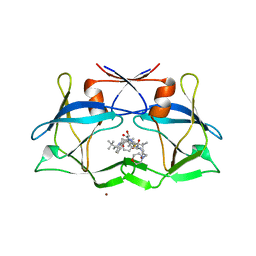 | | crystal structure of htlv protease complexed with the inhibitor KNI-10729 | | Descriptor: | N-{(1S,2S)-1-benzyl-3-[(4R)-5,5-dimethyl-4-{[(1R)-1,2,2-trimethylpropyl]carbamoyl}-1,3-thiazolidin-3-yl]-2-hydroxy-3-oxopropyl}-3-methyl-N~2~-{(2S)-2-[(morpholin-4-ylacetyl)amino]-2-phenylacetyl}-L-valinamide, Protease, ZINC ION | | Authors: | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | Deposit date: | 2010-01-25 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
8K93
 
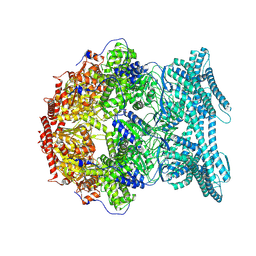 | | CryoEM structure of LonC protease S582A open hexamer with lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
3LK9
 
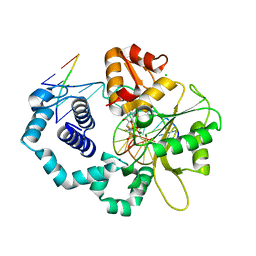 | | DNA polymerase beta with a gapped DNA substrate and dTMP(CF2)P(CF2)P | | Descriptor: | 5'-O-[(R)-{[(R)-[difluoro(phosphono)methyl](hydroxy)phosphoryl](difluoro)methyl}(hydroxy)phosphoryl]thymidine, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Zibinsky, M, Surya Prakash, G.K, Upton, T.G, Kashemirov, B.A, McKenna, C.E, Oertell, K, Goodman, M.F, Batra, V.K, Pedersen, L.C, Beard, W.A, Wilson, S.H. | | Deposit date: | 2010-01-27 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthesis and biological evaluation of fluorinated deoxynucleotide analogs based on bis-(difluoromethylene)triphosphoric acid.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6KVQ
 
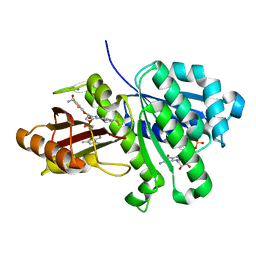 | | S. aureus FtsZ in complex with BOFP (compound 3) | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ferrer-Gonzalez, E, Fujita, J, Yoshizawa, T, Nelson, J.M, Pilch, A.J, Hillman, E, Ozawa, M, Kuroda, N, Parhi, A.K, LaVoie, E.J, Matsumura, H, Pilch, D.S. | | Deposit date: | 2019-09-05 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of a Fluorescent Probe for the Visualization of FtsZ in Clinically Important Gram-Positive and Gram-Negative Bacterial Pathogens.
Sci Rep, 9, 2019
|
|
