5XAG
 
 | | Crystal structure of tubulin-stathmin-TTL-Compound Z2 complex | | Descriptor: | (3~{R},4~{R})-3-(hydroxymethyl)-4-(4-methoxy-3-oxidanyl-phenyl)-1-(3,4,5-trimethoxyphenyl)azetidin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhang, H, Luo, C, Wang, Y. | | Deposit date: | 2017-03-12 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Design, synthesis, biological evaluation and cocrystal structures with tubulin of chiral beta-lactam bridged combretastatin A-4 analogues as potent antitumor agents
Eur J Med Chem, 144, 2017
|
|
3COQ
 
 | | Structural Basis for Dimerization in DNA Recognition by Gal4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*DAP*DCP*DCP*DGP*DGP*DAP*DGP*DGP*DAP*DCP*DAP*DGP*DTP*DCP*DCP*DTP*DCP*DCP*DGP*DG)-3'), DNA (5'-D(*DTP*DCP*DCP*DGP*DGP*DAP*DGP*DGP*DAP*DCP*DTP*DGP*DTP*DCP*DCP*DTP*DCP*DCP*DGP*DG)-3'), ... | | Authors: | Hong, M, Fitzgerald, M.X, Harper, S, Luo, C, Speicher, D.W. | | Deposit date: | 2008-03-29 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for dimerization in DNA recognition by gal4.
Structure, 16, 2008
|
|
7DPO
 
 | | Crystal Structure of BRD2(BD2)with Ligand ZB-BD-224 bound | | Descriptor: | 5-(1-naphthoyl)-11-methyl-8-((methylsulfonyl)methyl)-4,5-dihydro-2,3a1,5-triazadibenzo[cd,h]azulen-1(2H)-one, Bromodomain-containing protein 2 | | Authors: | Li, Z, Lu, T, Chen, P, Luo, C, Zhou, B. | | Deposit date: | 2020-12-21 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29994035 Å) | | Cite: | Structure Base Design of A new chemotype of Four-Cycle Compounds as Bromodomain and Extra-Terminal (BET) Inhibitors with The Second Bromodomain Bias and Highly Anti-inflammatory Potency
To Be Published
|
|
7DPN
 
 | | Crystal structure of BRD2(BD1)with ligand ZB-BD-224 bound | | Descriptor: | 5-(1-naphthoyl)-11-methyl-8-((methylsulfonyl)methyl)-4,5-dihydro-2,3a1,5-triazadibenzo[cd,h]azulen-1(2H)-one, Bromodomain-containing protein 2 | | Authors: | Li, Z, Lu, T, Chen, P, Luo, C, Zhou, B. | | Deposit date: | 2020-12-20 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79998839 Å) | | Cite: | Structure Base Design of A new chemotype of Four-Cycle Compounds as Bromodomain and Extra-Terminal (BET) Inhibitors with The Second Bromodomain Bias and Highly Anti-inflammatory Potency
To Be Published
|
|
7F5D
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-03 bound | | Descriptor: | 6-(1H-indol-5-yl)-N-methyl-2-methylsulfonyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.57150865 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7F5C
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-07 bound | | Descriptor: | 6-[1-[3-(dimethylamino)propyl]indol-5-yl]-2-methylsulfonyl-N-propyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65004492 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
7F5E
 
 | | Crystal structure of BPTF-BRD with ligand DC-BPi-11 bound | | Descriptor: | N,N-dimethyl-3-[5-(2-methylsulfonyl-7H-pyrrolo[2,3-d]pyrimidin-4-yl)indol-1-yl]propan-1-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B, Wang, J, Lin, H, Lu, W, Luo, C. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20017123 Å) | | Cite: | Discovery and Optimization of Small-Molecule Inhibitors for the BPTF Bromodomains Proteins
To Be Published
|
|
2IOO
 
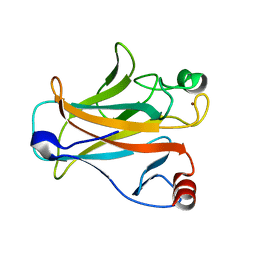 | | Crystal structure of the mouse p53 core domain | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Ho, W.C, Luo, C, Zhao, K, Chai, X, Fitzgerald, M.X, Marmorstein, R. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | High-resolution structure of the p53 core domain: implications for binding small-molecule stabilizing compounds.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2IOI
 
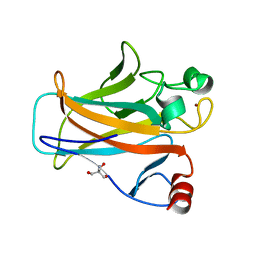 | | Crystal structure of the mouse p53 core domain at 1.55 A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular tumor antigen p53, ZINC ION | | Authors: | Ho, W.C, Luo, C, Zhao, K, Chai, X, Fitzgerald, M.X, Marmorstein, R. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-resolution structure of the p53 core domain: implications for binding small-molecule stabilizing compounds.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2IOM
 
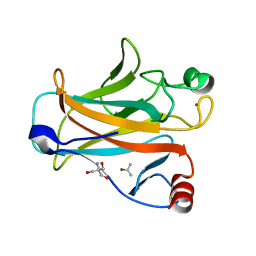 | | Mouse p53 core domain soaked with 2-propanol | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular tumor antigen p53, ISOPROPYL ALCOHOL, ... | | Authors: | Ho, W.C, Luo, C, Zhao, K, Chai, X, Fitzgerald, M.X, Marmorstein, R. | | Deposit date: | 2006-10-10 | | Release date: | 2006-12-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structure of the p53 core domain: implications for binding small-molecule stabilizing compounds.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
8JN8
 
 | |
8JN9
 
 | | Crystal structure of c-Src in complex with covalent inhibitor LW-Srci-8 | | Descriptor: | N-[(1R)-1-[3,5-bis(fluoranyl)phenyl]-2-(cyclopentylamino)-2-oxidanylidene-ethyl]-N-cyclopropyl-prop-2-enamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Zhang, H.M, Luo, C. | | Deposit date: | 2023-06-06 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.724 Å) | | Cite: | The crystal structure of c-Src in complex with covalent inhibitor LW-Srci-8
To Be Published
|
|
7KXT
 
 | | Crystal structure of human EED | | Descriptor: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Polycomb protein EED, UNKNOWN ATOM OR ION | | Authors: | Zhu, L, Dong, A, Du, D, Liu, Y, Luo, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Development of Small-Molecule PRC2 Inhibitors Targeting EZH2-EED Interaction.
J.Med.Chem., 64, 2021
|
|
6K05
 
 | | Crystal structure of BRD2(BD1)with ligand BY27 bound | | Descriptor: | (6~{R})-~{N}-(4-chlorophenyl)-1-methyl-8-(1-methylpyrazol-4-yl)-5,6-dihydro-4~{H}-[1,2,4]triazolo[4,3-a][1]benzazepin-6-amine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Lu, T, Lu, W, Chen, D, Zhao, Y, Luo, C. | | Deposit date: | 2019-05-05 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Discovery, structural insight, and bioactivities of BY27 as a selective inhibitor of the second bromodomains of BET proteins.
Eur.J.Med.Chem., 182, 2019
|
|
6K04
 
 | | Crystal structure of BRD2(BD2)with ligand BY27 bound | | Descriptor: | (6~{R})-~{N}-(4-chlorophenyl)-1-methyl-8-(1-methylpyrazol-4-yl)-5,6-dihydro-4~{H}-[1,2,4]triazolo[4,3-a][1]benzazepin-6-amine, Bromodomain-containing protein 2 | | Authors: | Lu, T, Lu, W, Chen, D, Zhao, Y, Luo, C. | | Deposit date: | 2019-05-05 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Discovery, structural insight, and bioactivities of BY27 as a selective inhibitor of the second bromodomains of BET proteins.
Eur.J.Med.Chem., 182, 2019
|
|
6KEE
 
 | | Crystal structure of BRD4 Bromodomain1 with an inhibitor | | Descriptor: | 6-[2-[2,4-bis(fluoranyl)phenoxy]-5-(methylsulfonylmethyl)pyridin-3-yl]-8-methyl-2H-pyrrolo[1,2-a]pyrazin-1-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Li, Z, Chen, S, Zhou, B, Luo, C. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12154651 Å) | | Cite: | Crystal structure of BRD4 Bromodomain1 with an inhibitor
To be published
|
|
6KEG
 
 | | BRD4 Bromodomain1 with an inhibitor | | Descriptor: | 6-[2-[2,4-bis(fluoranyl)phenoxy]-5-(ethylsulfonylmethyl)pyridin-3-yl]-8-methyl-4H-pyrrolo[1,2-a]pyrazin-1-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Li, Z, Chen, S, Zhou, B, Luo, C. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.232 Å) | | Cite: | BRD4 Bromodomain1 with an inhibitor
To Be Published
|
|
6KED
 
 | | BRD4 Bromodomain1 with an inhibitor | | Descriptor: | 6-[2-[2,4-bis(fluoranyl)phenoxy]-5-(methylsulfonylmethyl)pyridin-3-yl]-8-methyl-2H-pyrrolo[1,2-d][1,2,4]triazin-1-one, Bromodomain-containing protein 4 | | Authors: | Xiao, S, Li, Z, Chen, S, Zhou, B, Luo, C. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.548447 Å) | | Cite: | BRD4 Bromodomain1 with an inhibitor
To Be Published
|
|
6KEF
 
 | | BRD4 Bromodomain1 with an inhibitor | | Descriptor: | Bromodomain-containing protein 4, N-[3-(8-methyl-1-oxidanylidene-2H-pyrrolo[1,2-a]pyrazin-6-yl)-4-phenoxy-phenyl]methanesulfonamide | | Authors: | Xiao, S, Li, Z, Chen, S, Zhou, B, Luo, C. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.44467545 Å) | | Cite: | BRD4 Bromodomain1 with an inhibitor
To Be Published
|
|
8X73
 
 | | Crystal structure of Peroxiredoxin I in complex with compound 19-069 | | Descriptor: | Peroxiredoxin-1, methyl (2~{S})-2-[[(2~{R},4~{a}~{S},6~{a}~{R},6~{a}~{S},14~{a}~{S},14~{b}~{R})-2,4~{a},6~{a},6~{a},9,14~{a}-hexamethyl-10-oxidanyl-11-oxidanylidene-1,3,4,5,6,13,14,14~{b}-octahydropicen-2-yl]carbamoylamino]-3-oxidanyl-propanoate | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2023-11-22 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Discovery of a Novel Orally Bioavailable FLT3-PROTAC Degrader for Efficient Treatment of Acute Myeloid Leukemia and Overcoming Resistance of FLT3 Inhibitors.
J.Med.Chem., 67, 2024
|
|
8X71
 
 | | Crystal structure of Peroxiredoxin I in complex with compound 19-064 | | Descriptor: | Peroxiredoxin-1, methyl 3-[[(2~{R},4~{a}~{S},6~{a}~{R},6~{a}~{S},14~{a}~{S},14~{b}~{R})-2,4~{a},6~{a},6~{a},9,14~{a}-hexamethyl-10-oxidanyl-11-oxidanylidene-1,3,4,5,6,13,14,14~{b}-octahydropicen-2-yl]carbamoylamino]oxetane-3-carboxylate | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2023-11-22 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of a Novel Orally Bioavailable FLT3-PROTAC Degrader for Efficient Treatment of Acute Myeloid Leukemia and Overcoming Resistance of FLT3 Inhibitors.
J.Med.Chem., 67, 2024
|
|
3S04
 
 | |
3STA
 
 | | Crystal structure of ClpP in tetradecameric form from Staphylococcus aureus | | Descriptor: | ATP-dependent Clp protease proteolytic subunit | | Authors: | Zhang, J, Ye, F, Lan, L, Jiang, H, Luo, C, Yang, C.-G. | | Deposit date: | 2011-07-09 | | Release date: | 2011-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural switching of Staphylococcus aureus Clp protease: a key to understanding protease dynamics
J.Biol.Chem., 286, 2011
|
|
3ST9
 
 | | Crystal structure of ClpP in heptameric form from Staphylococcus aureus | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhang, J, Ye, F, Lan, L, Jiang, H, Luo, C, Yang, C.-G. | | Deposit date: | 2011-07-09 | | Release date: | 2011-09-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural switching of Staphylococcus aureus Clp protease: a key to understanding protease dynamics
J.Biol.Chem., 286, 2011
|
|
966C
 
 | | CRYSTAL STRUCTURE OF FIBROBLAST COLLAGENASE-1 COMPLEXED TO A DIPHENYL-ETHER SULPHONE BASED HYDROXAMIC ACID | | Descriptor: | CALCIUM ION, MMP-1, N-HYDROXY-2-[4-(4-PHENOXY-BENZENESULFONYL)-TETRAHYDRO-PYRAN-4-YL]-ACETAMIDE, ... | | Authors: | Lovejoy, B, Welch, A, Carr, S, Luong, C, Broka, C, Hendricks, R.T, Campbell, J, Walker, K, Martin, R, Van Wart, H, Browner, M.F. | | Deposit date: | 1998-08-07 | | Release date: | 1999-08-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of MMP-1 and -13 reveal the structural basis for selectivity of collagenase inhibitors.
Nat.Struct.Biol., 6, 1999
|
|
