3SJK
 
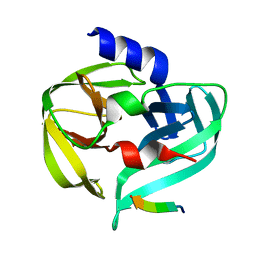 | | Crystal structure of the C147A mutant 3C from enterovirus 71 | | Descriptor: | 3C protease, KPVLRTATVQGPSLDF peptide | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
5ZKL
 
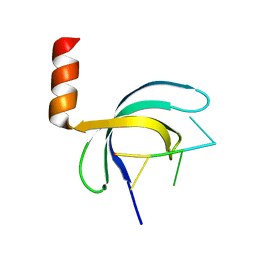 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA dT12 | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), SP_0782 | | Authors: | Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
5ZKM
 
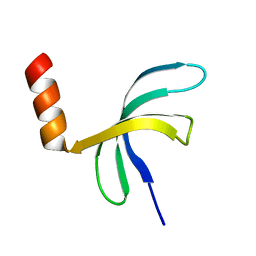 | | Crystal structure of Streptococcus pneumoniae SP_0782 (residues 7-79) in complex with single-stranded DNA TCTTCC | | Descriptor: | DNA (5'-D(P*TP*CP*TP*TP*CP*C)-3'), SP_0782 | | Authors: | Lu, G, Li, S, Zhu, J, Yang, Y, Gong, P. | | Deposit date: | 2018-03-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insight into the length-dependent binding of ssDNA by SP_0782 from Streptococcus pneumoniae, reveals a divergence in the DNA-binding interface of PC4-like proteins.
Nucleic Acids Res., 48, 2020
|
|
6A0P
 
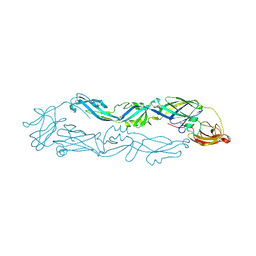 | | Crystal structure of Usutu virus envelope protein in the pre-fusion state | | Descriptor: | Envelope protein | | Authors: | Lu, G, Chen, Z, Ye, F, Lin, S, Yang, F, Cheng, Y. | | Deposit date: | 2018-06-06 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Usutu virus envelope protein in the pre-fusion state
Virol. J., 15, 2018
|
|
4MYV
 
 | | Free HSV-2 gD structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D | | Authors: | Lu, G, Zhang, N, Qi, J, Li, Y, Chen, Z, Zheng, C, Yan, J, Gao, G.F. | | Deposit date: | 2013-09-28 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of herpes simplex virus 2 gD bound to nectin-1 reveals a conserved mode of receptor recognition.
J.Virol., 88, 2014
|
|
4JL7
 
 | | Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First PDZ Domain of NHERF1 | | Descriptor: | CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Lu, G, Wu, Y, Jiang, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-03-12 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structural Insights into Neutrophilic Migration Revealed by the Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First PDZ Domain of NHERF1.
Plos One, 8, 2013
|
|
4K6M
 
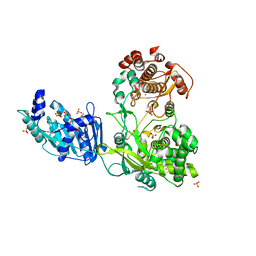 | | Crystal Structure of the full-length Japanese encephalitis virus NS5 | | Descriptor: | Polyprotein, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Lu, G, Gong, P. | | Deposit date: | 2013-04-16 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface
Plos Pathog., 9, 2013
|
|
4KR0
 
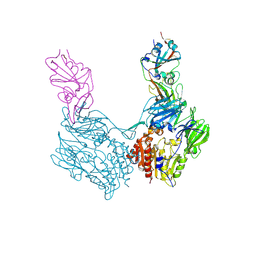 | | Complex structure of MERS-CoV spike RBD bound to CD26 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Lu, G, Hu, Y, Wang, Q, Qi, J, Gao, F, Li, Y, Zhang, Y, Zhang, W, Yuan, Y, Zhang, B, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-05-15 | | Release date: | 2013-07-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26.
Nature, 500, 2013
|
|
4N6X
 
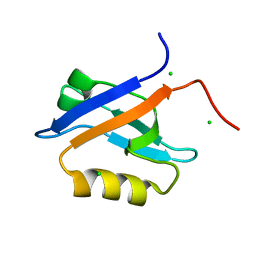 | | Crystal Structure of the Chemokine Receptor CXCR2 in Complex with the First PDZ Domain of NHERF1 | | Descriptor: | CHLORIDE ION, Na(+)/H(+) exchange regulatory cofactor NHE-RF1/Chemokine Receptor CXCR2 fusion protein | | Authors: | Lu, G, Wu, Y, Jiang, Y, Brunzelle, J, Sirinupong, N, Li, C, Yang, Z. | | Deposit date: | 2013-10-14 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.051 Å) | | Cite: | New Conformational State of NHERF1-CXCR2 Signaling Complex Captured by Crystal Lattice Trapping.
Plos One, 8, 2013
|
|
4MYW
 
 | | Structure of HSV-2 gD bound to nectin-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D, ... | | Authors: | Lu, G, Zhang, N, Qi, J, Li, Y, Chen, Z, Zheng, C, Yan, J, Gao, G.F. | | Deposit date: | 2013-09-28 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.189 Å) | | Cite: | Crystal structure of herpes simplex virus 2 gD bound to nectin-1 reveals a conserved mode of receptor recognition.
J.Virol., 88, 2014
|
|
4KQZ
 
 | | structure of the receptor binding domain (RBD) of MERS-CoV spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Lu, G, Hu, Y, Wang, Q, Qi, J, Gao, F, Li, Y, Zhang, Y, Zhang, W, Yuan, Y, Bao, J, Zhang, B, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2013-05-15 | | Release date: | 2013-07-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.514 Å) | | Cite: | Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26.
Nature, 500, 2013
|
|
6JHY
 
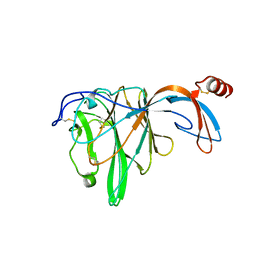 | |
4GJT
 
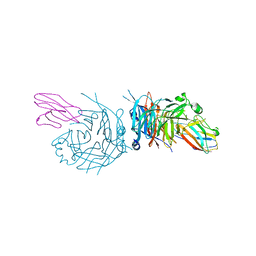 | | complex structure of nectin-4 bound to MV-H | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin glycoprotein, Poliovirus receptor-related protein 4 | | Authors: | Zhang, X, Lu, G, Qi, J, Li, Y, He, Y, Xu, X, Shi, J, Zhang, C, Yan, J, Gao, G.F. | | Deposit date: | 2012-08-10 | | Release date: | 2012-10-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1001 Å) | | Cite: | Structure of measles virus hemagglutinin bound to its epithelial receptor nectin-4
Nat.Struct.Mol.Biol., 20, 2013
|
|
8IIC
 
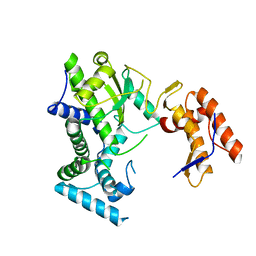 | |
8IIB
 
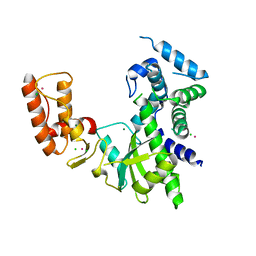 | | Crystal structure of Israeli acute paralysis virus RNA-dependent RNA polymerase delta85 mutant (residues 86-546) | | Descriptor: | CADMIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Fang, X, Lu, G, Hou, C, Gong, P. | | Deposit date: | 2023-02-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Unusual substructure conformations observed in crystal structures of a dicistrovirus RNA-dependent RNA polymerase suggest contribution of the N-terminal extension in proper folding.
Virol Sin, 38, 2023
|
|
9AYF
 
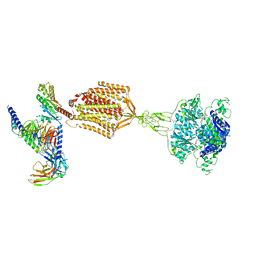 | | Structure of human calcium-sensing receptor in complex with Gi1 (miniGi1) protein in detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-07 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9AVG
 
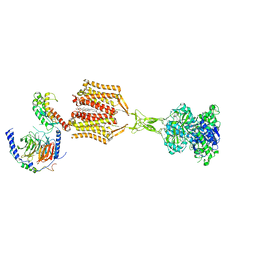 | | Structure of human calcium-sensing receptor in complex with chimeric Gs (miniGis) protein in nanodiscs | | Descriptor: | (19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-02 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9ASB
 
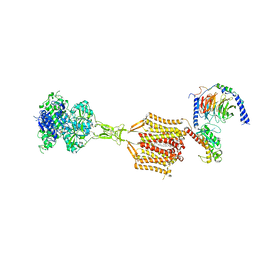 | | Structure of human calcium-sensing receptor in complex with chimeric Gq (miniGisq) protein in nanodiscs | | Descriptor: | (19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-02-24 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9AVL
 
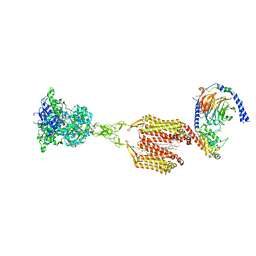 | | Structure of human calcium-sensing receptor in complex with Gi3 protein in nanodiscs | | Descriptor: | (19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacosan-19-yl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-04 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
9AXF
 
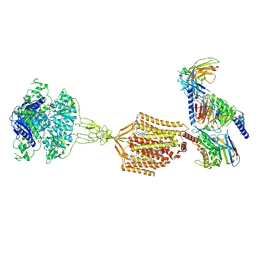 | | Structure of human calcium-sensing receptor in complex with chimeric Gq (miniGisq) protein in detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine, ... | | Authors: | Zuo, H, Park, J, Frangaj, A, Ye, J, Lu, G, Manning, J.J, Asher, W.B, Lu, Z, Hu, G, Wang, L, Mendez, J, Eng, E, Zhang, Z, Lin, X, Grasucci, R, Hendrickson, W.A, Clarke, O.B, Javitch, J.A, Conigrave, A.D, Fan, Q.R. | | Deposit date: | 2024-03-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Promiscuous G-protein activation by the calcium-sensing receptor.
Nature, 629, 2024
|
|
4EWJ
 
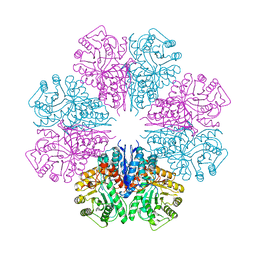 | | structure of the enloase from Streptococcus suis serotype 2 | | Descriptor: | Enolase 2 | | Authors: | Lu, Q, Lu, H, Qi, J, Lu, G, Gao, G.F. | | Deposit date: | 2012-04-27 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | An octamer of enolase from Streptococcus suis.
Protein Cell, 3, 2012
|
|
1HV4
 
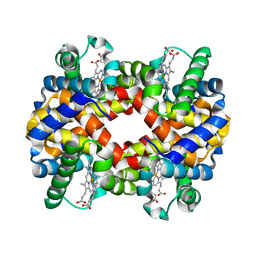 | | CRYSTAL STRUCTURE ANALYSIS OF BAR-HEAD GOOSE HEMOGLOBIN (DEOXY FORM) | | Descriptor: | HEMOGLOBIN ALPHA-A CHAIN, HEMOGLOBIN BETA CHAIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Liang, Y, Hua, Z, Liang, X, Xu, Q, Lu, G. | | Deposit date: | 2001-01-07 | | Release date: | 2001-01-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bar-headed goose hemoglobin in deoxy form: the allosteric mechanism of a hemoglobin species with high oxygen affinity.
J.Mol.Biol., 313, 2001
|
|
3FAN
 
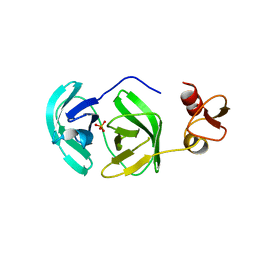 | | Crystal structure of chymotrypsin-like protease/proteinase (3CLSP/Nsp4) of porcine reproductive and respiratory syndrome virus (PRRSV) | | Descriptor: | Non-structural protein, PHOSPHATE ION | | Authors: | Gao, F, Peng, H, Tian, X, Lu, G, Liu, Y, Gao, G.F. | | Deposit date: | 2008-11-17 | | Release date: | 2009-09-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and cleavage specificity of the chymotrypsin-like serine protease (3CLSP/nsp4) of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV).
J.Mol.Biol., 392, 2009
|
|
3FAO
 
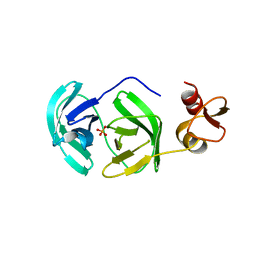 | | Crystal structure of S118A mutant 3CLSP of PRRSV | | Descriptor: | Non-structural protein, PHOSPHATE ION | | Authors: | Gao, F, Peng, H, Tian, X, Lu, G, Liu, Y, Gao, G.F. | | Deposit date: | 2008-11-17 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure and cleavage specificity of the chymotrypsin-like serine protease (3CLSP/nsp4) of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV).
J.Mol.Biol., 392, 2009
|
|
4NFB
 
 | | Structure of paired immunoglobulin-like type 2 receptor (PILR ) | | Descriptor: | Paired immunoglobulin-like type 2 receptor alpha | | Authors: | Lu, Q, Lu, G, Qi, J, Li, Y, Zhang, Y, Wang, H, Fan, Z, Yan, J, Gao, G. | | Deposit date: | 2013-10-31 | | Release date: | 2014-05-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PILR alpha and PILR beta have a siglec fold and provide the basis of binding to sialic acid
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
