7LNM
 
 | | Ornithine Aminotransferase (OAT) cocrystallized with its inactivator - (1S,3S)-3-amino-4-(difluoromethylene)cyclopentene-1-carboxylic acid | | Descriptor: | (1~{R},3~{S},4~{R})-3-methyl-4-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]cyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Butrin, A, Catlin, D, Zhu, W, Liu, D, Silverman, R. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Remarkable and Unexpected Mechanism for ( S )-3-Amino-4-(difluoromethylenyl)cyclohex-1-ene-1-carboxylic Acid as a Selective Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
7LON
 
 | | Ornithine Aminotransferase (OAT) cocrystallized with its inactivator - (1S,3S)-3-amino-4-(difluoromethylene)cyclohexene-1-carboxylic acid | | Descriptor: | (1R,3S,4R)-3-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-4-methylcyclohexane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial, ... | | Authors: | Butrin, A, Zhu, W, Liu, D, Silverman, R. | | Deposit date: | 2021-02-10 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Remarkable and Unexpected Mechanism for ( S )-3-Amino-4-(difluoromethylenyl)cyclohex-1-ene-1-carboxylic Acid as a Selective Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
7LOM
 
 | | Ornithine Aminotransferase (OAT) soaked with its inactivator - (1S,3S)-3-amino-4-(difluoromethylene)cyclohexene-1-carboxylic acid | | Descriptor: | (3~{S},4~{S})-4-methyl-3-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]cyclohexene-1-carboxylic acid, (4~{R})-4-(fluoranylmethyl)-3-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]cyclohexene-1-carboxylic acid, Ornithine aminotransferase, ... | | Authors: | Butrin, A, Zhu, W, Liu, D, Silverman, R. | | Deposit date: | 2021-02-10 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Remarkable and Unexpected Mechanism for ( S )-3-Amino-4-(difluoromethylenyl)cyclohex-1-ene-1-carboxylic Acid as a Selective Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
7M31
 
 | | Dihydropyrimidine Dehydrogenase (DPD) C671S Mutant Soaked with Thymine and NADPH Anaerobically | | Descriptor: | Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Butrin, A, Beaupre, B, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Perturbing the Movement of Hydrogens to Delineate and Assign Events in the Reductive Activation and Turnover of Porcine Dihydropyrimidine Dehydrogenase.
Biochemistry, 60, 2021
|
|
5UBQ
 
 | | Cryo-EM structure of ciliary microtubule doublet | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ichikawa, M, Liu, D, Kastritis, P.L, Basu, K, Bui, K.H. | | Deposit date: | 2016-12-21 | | Release date: | 2017-05-10 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Subnanometre-resolution structure of the doublet microtubule reveals new classes of microtubule-associated proteins.
Nat Commun, 8, 2017
|
|
7LJT
 
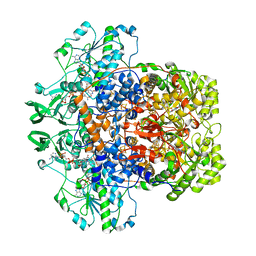 | | Porcine Dihydropyrimidine Dehydrogenase (DPD) soaked with 5-Ethynyluracil (5EU), NADPH - 20 minutes | | Descriptor: | 5-ethynylpyrimidine-2,4(1H,3H)-dione, Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN MONONUCLEOTIDE, ... | | Authors: | Butrin, A, Forouzesh, D, Beaupre, B, Wawrzak, Z, Liu, D, Moran, G. | | Deposit date: | 2021-01-30 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Interaction of Porcine Dihydropyrimidine Dehydrogenase with the Chemotherapy Sensitizer: 5-Ethynyluracil.
Biochemistry, 60, 2021
|
|
7LJS
 
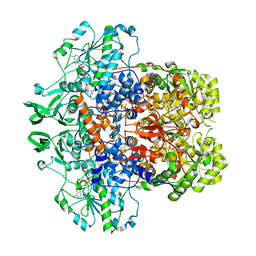 | | Porcine Dihydropyrimidine dehydrogenase (DPD) complexed with 5-Ethynyluracil (5EU) - Open Form | | Descriptor: | 5-ethynylpyrimidine-2,4(1H,3H)-dione, Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN MONONUCLEOTIDE, ... | | Authors: | Butrin, A, Forouzesh, D, Beaupre, B, Wawrzak, Z, Liu, D, Moran, G. | | Deposit date: | 2021-01-30 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Interaction of Porcine Dihydropyrimidine Dehydrogenase with the Chemotherapy Sensitizer: 5-Ethynyluracil.
Biochemistry, 60, 2021
|
|
4PVG
 
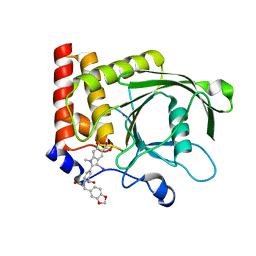 | |
5UCY
 
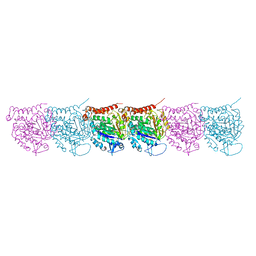 | | Cryo-EM map of protofilament of microtubule doublet | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ichikawa, M, Liu, D, Kastritis, P.L, Basu, K, Bui, K.H. | | Deposit date: | 2016-12-22 | | Release date: | 2017-05-10 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Subnanometre-resolution structure of the doublet microtubule reveals new classes of microtubule-associated proteins.
Nat Commun, 8, 2017
|
|
7LK0
 
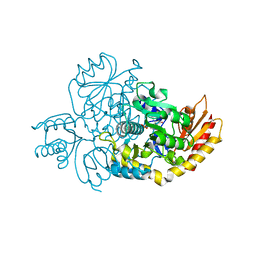 | | Ornithine Aminotransferase (OAT) cocrystallized with its potent inhibitor - (S)-3-amino-4,4-difluorocyclopent-1-enecarboxylic acid (SS-1-148) | | Descriptor: | (1R,3S)-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-oxocyclopentane-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Butrin, A, Shen, S, Liu, D, Silverman, R. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Turnover and Inactivation Mechanisms for ( S )-3-Amino-4,4-difluorocyclopent-1-enecarboxylic Acid, a Selective Mechanism-Based Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
1R8T
 
 | | Solution structures of high affinity miniprotein ligands to Streptavidin | | Descriptor: | MP1 | | Authors: | Luo, J, Mukherjee, M, Fan, X, Yang, H, Liu, D, Khan, R, White, M, Fox, R.O. | | Deposit date: | 2003-10-28 | | Release date: | 2005-02-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure-based design of high affinity miniprotein ligands
To be Published
|
|
7LK1
 
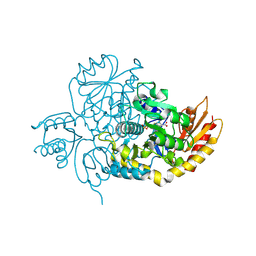 | | Ornithine Aminotransferase (OAT) with its potent inhibitor - (S)-3-amino-4,4-difluorocyclopent-1-enecarboxylic acid (SS-1-148) - 1 Hour Soaking | | Descriptor: | (1R,4R)-4-fluoro-3-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]cyclopent-2-ene-1-carboxylic acid, Ornithine aminotransferase, mitochondrial | | Authors: | Butrin, A, Shen, S, Liu, D, Silverman, R. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Turnover and Inactivation Mechanisms for ( S )-3-Amino-4,4-difluorocyclopent-1-enecarboxylic Acid, a Selective Mechanism-Based Inactivator of Human Ornithine Aminotransferase.
J.Am.Chem.Soc., 143, 2021
|
|
6BY7
 
 | | Folding DNA into a lipid-conjugated nano-barrel for controlled reconstitution of membrane proteins | | Descriptor: | DNA (26-MER), DNA (27-MER), DNA (29-MER), ... | | Authors: | Dong, Y, Chen, S, Zhang, S, Sodroski, J, Yang, Z, Liu, D, Mao, Y. | | Deposit date: | 2017-12-20 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Folding DNA into a Lipid-Conjugated Nanobarrel for Controlled Reconstitution of Membrane Proteins.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7M7C
 
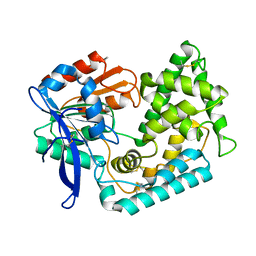 | | Crystal Structure of Hip1 (Rv2224c) mutant - T466A/S228DHA (dehydroalanine) | | Descriptor: | Carboxylesterase A | | Authors: | Naffin-Olivos, J.L, Daab, A, Goldfarb, N.E, Doran, M.H, Baikovitz, J, Liu, D, Sun, S, White, A, Dunn, B.M, Rengarajan, J, Petsko, G.A, Ringe, D. | | Deposit date: | 2021-03-27 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitors and Inactivators of Mycobacterium tuberculosis serine protease Hip1 (Rv2224c)
To Be Published
|
|
7JX9
 
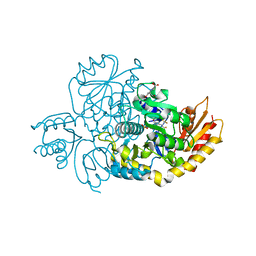 | | The crystal structure of human ornithine aminotransferase with an intermediate bound during inactivation by (1S,3S)-3-amino-4-(hexafluoropropan-2-ylidenyl)-cyclopentane-1-carboxylic acid. | | Descriptor: | (1S,3S,4S)-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-(1,1,3,3,3-pentafluoroprop-1-en-2-yl)cyclopentane-1-carboxylic acid, N-[1,3-dihydroxy-2-(hydroxymethyl)propan-2-yl]glycine, Ornithine aminotransferase, ... | | Authors: | Butrin, A, Beaupre, B, Shen, S, Silverman, R.B, Moran, G, Liu, D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and Kinetic Analyses Reveal the Dual Inhibition Modes of Ornithine Aminotransferase by (1 S ,3 S )-3-Amino-4-(hexafluoropropan-2-ylidenyl)-cyclopentane-1-carboxylic Acid (BCF 3 ).
Acs Chem.Biol., 16, 2021
|
|
3CVA
 
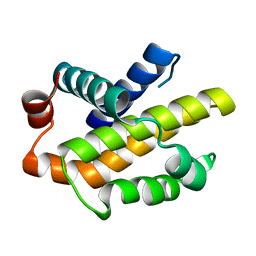 | | Human Bcl-xL containing a Trp to Ala mutation at position 137 | | Descriptor: | Apoptosis regulator Bcl-X | | Authors: | Feng, Y, Zhang, L, Hu, T, Shen, X, Chen, K, Jiang, H, Liu, D. | | Deposit date: | 2008-04-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A conserved hydrophobic core at Bcl-x(L) mediates its structural stability and binding affinity with BH3-domain peptide of pro-apoptotic protein
Arch.Biochem.Biophys., 484, 2009
|
|
4FM4
 
 | | Wild Type Fe-type Nitrile Hydratase from Comamonas testosteroni Ni1 | | Descriptor: | FE (III) ION, Nitrile hydratase alpha subunit, Nitrile hydratase beta subunit, ... | | Authors: | Kuhn, M.L, Martinez, S, Gumataotao, N, Bornscheuer, U, Liu, D, Holz, R.C. | | Deposit date: | 2012-06-15 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.384 Å) | | Cite: | The Fe-type nitrile hydratase from Comamonas testosteroni Ni1 does not require an activator accessory protein for expression in Escherichia coli.
Biochem.Biophys.Res.Commun., 424, 2012
|
|
6BYZ
 
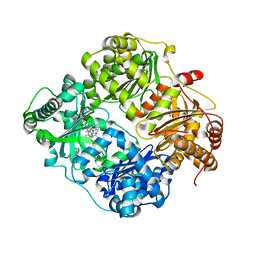 | | Structure of Cysteine-free Human Insulin-Degrading Enzyme in complex with Substrate-selective Macrocyclic Inhibitor 37 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ALA-ALA-ALA, Insulin-degrading enzyme, ... | | Authors: | Tan, G.A, Seeliger, M.A, Maianti, J.P, Liu, D.R. | | Deposit date: | 2017-12-21 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.95624852 Å) | | Cite: | Substrate-selective inhibitors that reprogram the activity of insulin-degrading enzyme.
Nat.Chem.Biol., 15, 2019
|
|
6B6G
 
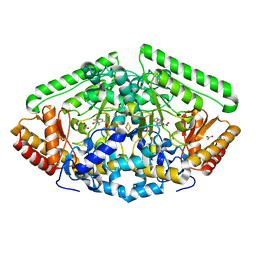 | | Crystal Structure of GABA Aminotransferase bound to (S)-3-Amino-4-(difluoromethylenyl)cyclopent-1-ene-1-carboxylic acid, an Potent Inactivatorfor the Treatment of Addiction | | Descriptor: | (3R,4E)-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]cyclopent-1-ene-1,3-dicarboxylic acid, 4-aminobutyrate aminotransferase, mitochondrial, ... | | Authors: | Mascarenhas, R, Juncosa, J.I, Takaya, K, Le, L.V, Moschitto, M.J, Silverman, R.B, Liu, D. | | Deposit date: | 2017-10-02 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design and Mechanism of (S)-3-Amino-4-(difluoromethylenyl)cyclopent-1-ene-1-carboxylic Acid, a Highly Potent gamma-Aminobutyric Acid Aminotransferase Inactivator for the Treatment of Addiction.
J. Am. Chem. Soc., 140, 2018
|
|
4IRW
 
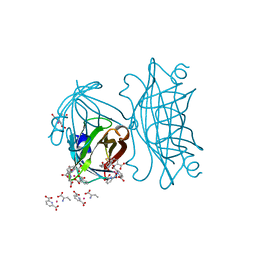 | | Co-crystallization of streptavidin-biotin complex with a lanthanide-ligand complex gives rise to a novel crystal form | | Descriptor: | BIOTIN, PYRIDINE-2,6-DICARBOXYLIC ACID, SODIUM ION, ... | | Authors: | Bandara, R.A.M.S.S, Liu, D.Q, Hindupur, A, Tesh, K.F, Fox, R.O. | | Deposit date: | 2013-01-15 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Co-crystallization of streptavidin-biotin complex with a lanthanide-ligand complex gives rise to a novel crystal form
To be Published
|
|
7JH4
 
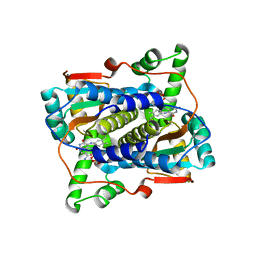 | | Crystal structure of NAD(P)H-flavin oxidoreductase (NfoR) from S. aureus complexed with reduced FMN and NAD+ | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, NAD(P)H-dependent oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zheng, Y, O'Neill, A.G, Beaupre, B.A, Liu, D, Moran, G.R. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | NfoR: Chromate Reductase or Flavin Mononucleotide Reductase?
Appl.Environ.Microbiol., 86, 2020
|
|
3U4W
 
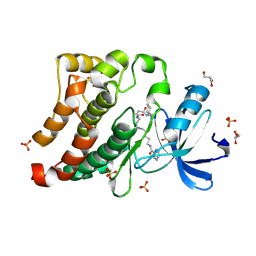 | | Src in complex with DNA-templated macrocyclic inhibitor MC4b | | Descriptor: | GLYCEROL, Proto-oncogene tyrosine-protein kinase Src, SULFATE ION, ... | | Authors: | Seeliger, M.A, Liu, D.R, Georghiou, G, Kleiner, R.E, Pulkoski-Gross, M. | | Deposit date: | 2011-10-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Highly specific, bisubstrate-competitive Src inhibitors from DNA-templated macrocycles.
Nat.Chem.Biol., 8, 2012
|
|
3U51
 
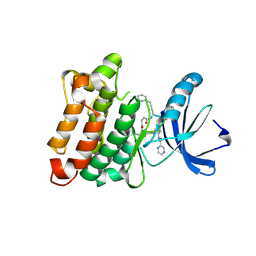 | | Src in complex with DNA-templated macrocyclic inhibitor MC1 | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, macrocyclic inhibitor MC1 | | Authors: | Seeliger, M.A, Liu, D.R, Georghiou, G, Kleiner, R.E, Pulkoski-Gross, M. | | Deposit date: | 2011-10-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | Highly specific, bisubstrate-competitive Src inhibitors from DNA-templated macrocycles.
Nat.Chem.Biol., 8, 2012
|
|
4MGR
 
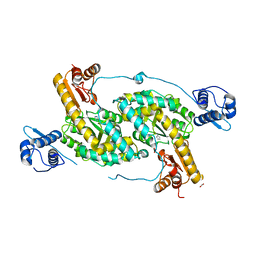 | | The crystal structure of Bacillus subtilis GabR, an autorepressor and PLP- and GABA-dependent transcriptional activator of gabT | | Descriptor: | ACETATE ION, HTH-type transcriptional regulatory protein GabR, IMIDAZOLE, ... | | Authors: | Wu, R, Edayathumangalam, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-08-28 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4N0B
 
 | | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of GabT | | Descriptor: | ACETYL GROUP, CALCIUM ION, HTH-type transcriptional regulatory protein GabR, ... | | Authors: | Edayathumangalam, R, Wu, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-10-01 | | Release date: | 2013-10-30 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
