6LU8
 
 | | Cryo-EM structure of a human pre-60S ribosomal subunit - state A | | 分子名称: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | 登録日 | 2020-01-26 | | 公開日 | 2020-08-26 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
6LQM
 
 | | Cryo-EM structure of a pre-60S ribosomal subunit - state C | | 分子名称: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | 登録日 | 2020-01-14 | | 公開日 | 2020-08-26 | | 実験手法 | ELECTRON MICROSCOPY (3.09 Å) | | 主引用文献 | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
6LSR
 
 | | Cryo-EM structure of a pre-60S ribosomal subunit - state B | | 分子名称: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | 著者 | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | 登録日 | 2020-01-20 | | 公開日 | 2020-08-26 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
6LSS
 
 | | Cryo-EM structure of a pre-60S ribosomal subunit - state preA | | 分子名称: | 28S rRNA, 5S rRNA, 60S ribosomal protein L11, ... | | 著者 | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | 登録日 | 2020-01-20 | | 公開日 | 2020-08-26 | | 実験手法 | ELECTRON MICROSCOPY (3.23 Å) | | 主引用文献 | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
1HV4
 
 | | CRYSTAL STRUCTURE ANALYSIS OF BAR-HEAD GOOSE HEMOGLOBIN (DEOXY FORM) | | 分子名称: | HEMOGLOBIN ALPHA-A CHAIN, HEMOGLOBIN BETA CHAIN, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Liang, Y, Hua, Z, Liang, X, Xu, Q, Lu, G. | | 登録日 | 2001-01-07 | | 公開日 | 2001-01-17 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The crystal structure of bar-headed goose hemoglobin in deoxy form: the allosteric mechanism of a hemoglobin species with high oxygen affinity.
J.Mol.Biol., 313, 2001
|
|
6YCS
 
 | | Human Transcription Cofactor PC4 DNA-binding domain in complex with full phosphorothioate 5-10-5 2'-O-methyl DNA gapmer antisense oligonucleotide. | | 分子名称: | DNA (5'-D(P*(OKQ))-D(P*(OKT))-R(P*(RFJ))-D(*(OKQ)P*(OKT)P*(AS)P*(GS)P*(OKN)P*(OKN)P*(PST)P*(OKN)P*(PST)P*(GS)P*(GS)P*(AS)P*(OKT)P*(OKT))-3'), PC4 protein, SODIUM ION, ... | | 著者 | Hyjek-Skladanowska, M, Vickers, T.A, Napiorkowska, A, Anderson, B, Tanowitz, M, Crooke, S.T, Liang, X, Seth, P.P, Nowotny, M. | | 登録日 | 2020-03-19 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Origins of the Increased Affinity of Phosphorothioate-Modified Therapeutic Nucleic Acids for Proteins.
J.Am.Chem.Soc., 142, 2020
|
|
1GWZ
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THE PROTEIN TYROSINE PHOSPHATASE SHP-1 | | 分子名称: | SHP-1 | | 著者 | Yang, J, Liang, X, Niu, T, Meng, W, Zhao, Z, Zhou, G.W. | | 登録日 | 1998-08-22 | | 公開日 | 1999-08-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of the catalytic domain of protein-tyrosine phosphatase SHP-1.
J.Biol.Chem., 273, 1998
|
|
1PS9
 
 | | The Crystal Structure and Reaction Mechanism of E. coli 2,4-Dienoyl CoA Reductase | | 分子名称: | 2,4-dienoyl-CoA reductase, 5-MERCAPTOETHANOL-2-DECENOYL-COENZYME A, CHLORIDE ION, ... | | 著者 | Hubbard, P.A, Liang, X, Schulz, H, Kim, J.J. | | 登録日 | 2003-06-20 | | 公開日 | 2003-09-30 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The crystal structure and reaction mechanism of Escherichia coli 2,4-dienoyl-CoA reductase
J.Biol.Chem., 278, 2003
|
|
3QKW
 
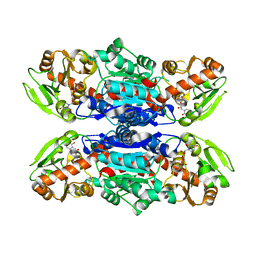 | | Structure of Streptococcus parasangunini Gtf3 glycosyltransferase | | 分子名称: | Nucleotide sugar synthetase-like protein, URIDINE-5'-DIPHOSPHATE | | 著者 | Zhu, F, Erlandsen, H, Huang, Y, Ding, L, Zhou, M, Liang, X, Ma, J.-B, Wu, H. | | 登録日 | 2011-02-01 | | 公開日 | 2011-06-08 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.287 Å) | | 主引用文献 | Structural and Functional Analysis of a New Subfamily of Glycosyltransferases Required for Glycosylation of Serine-rich Streptococcal Adhesins.
J.Biol.Chem., 286, 2011
|
|
7CFM
 
 | | Cryo-EM structure of the P395-bound GPBAR-Gs complex | | 分子名称: | 2-(ethylamino)-6-[3-(4-propan-2-ylphenyl)propanoyl]-7,8-dihydro-5H-pyrido[4,3-d]pyrimidine-4-carboxamide, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | 著者 | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | 登録日 | 2020-06-27 | | 公開日 | 2020-09-09 | | 最終更新日 | 2020-12-02 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
7CFN
 
 | | Cryo-EM structure of the INT-777-bound GPBAR-Gs complex | | 分子名称: | (2S,4R)-4-[(3R,5S,6R,7R,8R,9S,10S,12S,13R,14S,17R)-6-ethyl-10,13-dimethyl-3,7,12-tris(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2-methyl-pentanoic acid, CHOLESTEROL, G-protein coupled bile acid receptor 1, ... | | 著者 | Yang, F, Mao, C, Guo, L, Lin, J, Ming, Q, Xiao, P, Wu, X, Shen, Q, Guo, S, Shen, D, Lu, R, Zhang, L, Huang, S, Ping, Y, Zhang, C, Ma, C, Zhang, K, Liang, X, Shen, Y, Nan, F, Yi, F, Luca, V, Zhou, J, Jiang, C, Sun, J, Xie, X, Yu, X, Zhang, Y. | | 登録日 | 2020-06-27 | | 公開日 | 2020-09-09 | | 最終更新日 | 2021-04-07 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of GPBAR activation and bile acid recognition.
Nature, 587, 2020
|
|
5CHL
 
 | | Structural basis of H2A.Z recognition by YL1 histone chaperone component of SRCAP/SWR1 chromatin remodeling complex | | 分子名称: | Histone H2A.Z, Vacuolar protein sorting-associated protein 72 homolog | | 著者 | Shan, S, Liang, X, Pan, L, Wu, C, Zhou, Z. | | 登録日 | 2015-07-10 | | 公開日 | 2016-03-09 | | 最終更新日 | 2017-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.892 Å) | | 主引用文献 | Structural basis of H2A.Z recognition by SRCAP chromatin-remodeling subunit YL1
Nat.Struct.Mol.Biol., 23, 2016
|
|
1YIK
 
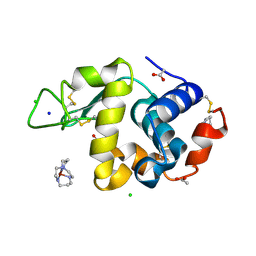 | | Structure of Hen egg white lysozyme soaked with Cu-cyclam | | 分子名称: | 1,4,8,11-TETRAAZA-CYCLOTETRADECANE CU(II), ACETATE ION, CHLORIDE ION, ... | | 著者 | Hunter, T.M, McNae, I.W, Liang, X, Bella, J, Parsons, S, Walkinshaw, M.D, Sadler, P.J. | | 登録日 | 2005-01-12 | | 公開日 | 2005-02-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Protein recognition of macrocycles: binding of anti-HIV metallocyclams to lysozyme
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1YIL
 
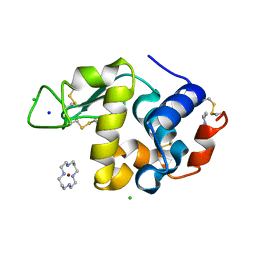 | | Structure of Hen egg white lysozyme soaked with Cu2-Xylylbicyclam | | 分子名称: | 1,1'-[1,4-PHENYLENEBIS(METHYLENE)]BIS[1,4,8,11-TETRAAZA-CYCLOTETRADECANE]CU(II)2, CHLORIDE ION, Lysozyme C, ... | | 著者 | Hunter, T.M, McNae, I.W, Liang, X, Bella, J, Parsons, S, Walkinshaw, M.D, Sadler, P.J. | | 登録日 | 2005-01-12 | | 公開日 | 2005-02-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Protein recognition of macrocycles: binding of anti-HIV metallocyclams to lysozyme
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6AKO
 
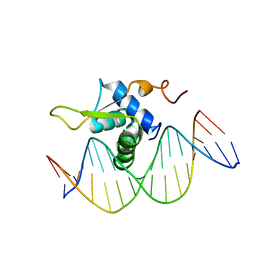 | | Crystal Structure of FOXC2 DBD Bound to DBE2 DNA | | 分子名称: | DNA (5'-D(CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), Forkhead box protein C2, ... | | 著者 | Chen, X, Wei, H, Li, J, Liang, X, Dai, S, Jiang, L, Guo, M, Chen, Y. | | 登録日 | 2018-09-03 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.396 Å) | | 主引用文献 | Structural basis for DNA recognition by FOXC2.
Nucleic Acids Res., 47, 2019
|
|
6AKP
 
 | | Crystal Structural of FOXC2 DNA binding domain bound to PC promoter | | 分子名称: | DNA (5'-D(AP*CP*AP*CP*AP*AP*AP*TP*AP*TP*TP*TP*GP*TP*GP*T)-3'), Forkhead box protein C2, MAGNESIUM ION | | 著者 | Chen, X, Wei, H, Li, J, Liang, X, Dai, S, Jiang, L, Guo, M, Chen, Y. | | 登録日 | 2018-09-03 | | 公開日 | 2019-02-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.323 Å) | | 主引用文献 | Structural basis for DNA recognition by FOXC2.
Nucleic Acids Res., 47, 2019
|
|
4N9E
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | 分子名称: | 1,2-ETHANEDIOL, 1-[(1-benzoylpiperidin-4-yl)methyl]-N-(pyridin-3-yl)-1H-benzimidazole-5-carboxamide, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | 著者 | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | 登録日 | 2013-10-20 | | 公開日 | 2014-02-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
1YP1
 
 | | Crystal structure of a non-hemorrhagic fibrin(ogen)olytic metalloproteinase from venom of Agkistrodon acutus | | 分子名称: | FII, KNL, ZINC ION | | 著者 | Lou, Z, Hou, J, Chen, J, Liang, X, Qiu, P, Liu, Y, Li, M, Rao, Z. | | 登録日 | 2005-01-28 | | 公開日 | 2006-01-17 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of a non-hemorrhagic fibrin(ogen)olytic metalloproteinase complexed with a novel natural tri-peptide inhibitor from venom of Agkistrodon acutus
J.Struct.Biol., 152, 2005
|
|
4N9B
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | 分子名称: | 1-methyl-N-(pyridin-3-yl)-1H-pyrazole-5-carboxamide, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | 著者 | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhai, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | 登録日 | 2013-10-20 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.859 Å) | | 主引用文献 | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N9D
 
 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | 分子名称: | 1,2-ETHANEDIOL, 4-({[(4-tert-butylphenyl)sulfonyl]amino}methyl)-N-(pyridin-3-yl)benzamide, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | 登録日 | 2013-10-20 | | 公開日 | 2014-02-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2B3O
 
 | | Crystal structure of human tyrosine phosphatase SHP-1 | | 分子名称: | Tyrosine-protein phosphatase, non-receptor type 6 | | 著者 | Yang, J, Liu, L, He, D, Song, X, Liang, X, Zhao, Z.J, Zhou, G.W. | | 登録日 | 2005-09-20 | | 公開日 | 2005-10-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of human protein-tyrosine phosphatase SHP-1.
J.Biol.Chem., 278, 2003
|
|
4N9C
 
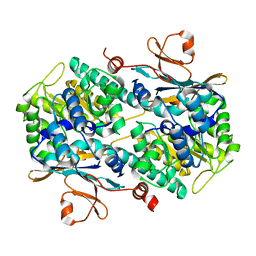 | | Fragment-based Design of 3-Aminopyridine-derived Amides as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | 分子名称: | 5-nitro-1H-benzimidazole, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | 著者 | Dragovich, P.S, Zhao, G, Baumeister, T, Bravo, B, Giannetti, A.M, Ho, Y, Hua, R, Li, G, Liang, X, O'Brien, T, Skelton, N.J, Wang, C, Zhao, Q, Oh, A, Wang, W, Wang, Y, Xiao, Y, Yuen, P, Zak, M, Zheng, X. | | 登録日 | 2013-10-20 | | 公開日 | 2014-02-19 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.751 Å) | | 主引用文献 | Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4NFT
 
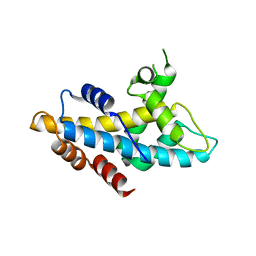 | | Crystal structure of human lnkH2B-h2A.Z-Anp32e | | 分子名称: | Acidic leucine-rich nuclear phosphoprotein 32 family member E, Histone H2B type 2-E, Histone H2A.Z | | 著者 | Shan, S, Pan, L, Mao, Z, Wang, W, Sun, J, Dong, Q, Liang, X, Ding, X, Chen, S, Dai, L, Zhang, Z, Zhu, B, Zhou, Z. | | 登録日 | 2013-11-01 | | 公開日 | 2014-04-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Anp32e, a higher eukaryotic histone chaperone directs preferential recognition for H2A.Z
Cell Res., 24, 2014
|
|
7CJT
 
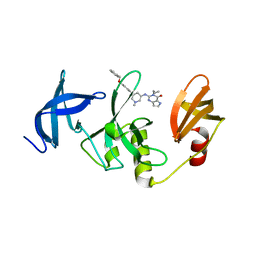 | | Crystal Structure of SETDB1 Tudor domain in complexed with (R,R)-59 | | 分子名称: | 2-[[(3~{R},5~{R})-1-methyl-5-(4-phenylmethoxyphenyl)piperidin-3-yl]amino]-3-prop-2-enyl-5~{H}-pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | 著者 | Guo, Y.P, Liang, X, Mao, X, Wu, C, Luyi, H, Yang, S. | | 登録日 | 2020-07-13 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.474 Å) | | 主引用文献 | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7CAJ
 
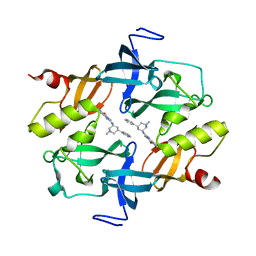 | | Crystal structure of SETDB1 Tudor domain in complexed with Compound 2. | | 分子名称: | 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-5H-pyrrolo[3,2-d]pyrimidin-4-one, Histone-lysine N-methyltransferase SETDB1 | | 著者 | Guo, Y.P, Liang, X, Xin, M, Luyi, H, Chengyong, W, Yang, S.Y. | | 登録日 | 2020-06-08 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.198 Å) | | 主引用文献 | Structure-Guided Discovery of a Potent and Selective Cell-Active Inhibitor of SETDB1 Tudor Domain.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
