4WPE
 
 | | Crystal Structure of Hof1p F-BAR domain | | Descriptor: | Cytokinesis protein 2 | | Authors: | Lemmon, M.A, Moravcevic, K. | | Deposit date: | 2014-10-17 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparison of Saccharomyces cerevisiae F-BAR Domain Structures Reveals a Conserved Inositol Phosphate Binding Site.
Structure, 23, 2015
|
|
4WPC
 
 | |
7TVD
 
 | |
6VG3
 
 | |
5JEB
 
 | |
1MAI
 
 | | STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM PHOSPHOLIPASE C DELTA IN COMPLEX WITH INOSITOL TRISPHOSPHATE | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, PHOSPHOLIPASE C DELTA-1 | | Authors: | Ferguson, K.M, Lemmon, M.A, Schlessinger, J, Sigler, P.B. | | Deposit date: | 1996-05-23 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain.
Cell(Cambridge,Mass.), 83, 1995
|
|
1NQL
 
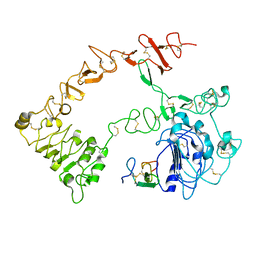 | |
1DYN
 
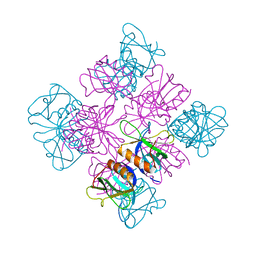 | | CRYSTAL STRUCTURE AT 2.2 ANGSTROMS RESOLUTION OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM HUMAN DYNAMIN | | Descriptor: | DYNAMIN | | Authors: | Ferguson, K.M, Lemmon, M.A, Schlessinger, J, Sigler, P.B. | | Deposit date: | 1994-12-21 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure at 2.2 A resolution of the pleckstrin homology domain from human dynamin.
Cell(Cambridge,Mass.), 79, 1994
|
|
1FAO
 
 | | STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM DAPP1/PHISH IN COMPLEX WITH INOSITOL 1,3,4,5-TETRAKISPHOSPHATE | | Descriptor: | DUAL ADAPTOR OF PHOSPHOTYROSINE AND 3-PHOSPHOINOSITIDES, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-07-13 | | Release date: | 2000-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains.
Mol.Cell, 6, 2000
|
|
1FHX
 
 | | Structure of the pleckstrin homology domain from GRP1 in complex with inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | GUANINE NUCLEOTIDE EXCHANGE FACTOR AND INTEGRIN BINDING PROTEIN HOMOLOG GRP1, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, SULFATE ION | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-08-02 | | Release date: | 2000-08-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains
Mol.Cell, 6, 2000
|
|
1FB8
 
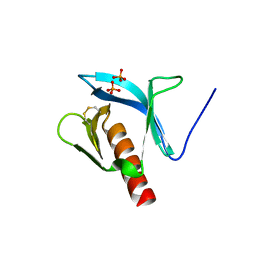 | | STRUCTURE OF THE PLECKSTRIN HOMOLOGY DOMAIN FROM DAPP1/PHISH | | Descriptor: | DUAL ADAPTOR OF PHOSPHOTYROSINE AND 3-PHOSPHOINOSITIDES, PHOSPHATE ION | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-07-14 | | Release date: | 2000-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains.
Mol.Cell, 6, 2000
|
|
1FHW
 
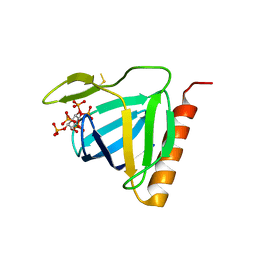 | | Structure of the pleckstrin homology domain from GRP1 in complex with inositol(1,3,4,5,6)pentakisphosphate | | Descriptor: | GUANINE NUCLEOTIDE EXCHANGE FACTOR AND INTEGRIN BINDING PROTEIN HOMOLOG GRP1, INOSITOL-(1,3,4,5,6)-PENTAKISPHOSPHATE, SULFATE ION | | Authors: | Ferguson, K.M, Kavran, J.M, Sankaran, V.G, Fournier, E, Isakoff, S.J, Skolnik, E.Y, Lemmon, M.A. | | Deposit date: | 2000-08-02 | | Release date: | 2000-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains
Mol.Cell, 6, 2000
|
|
1IHK
 
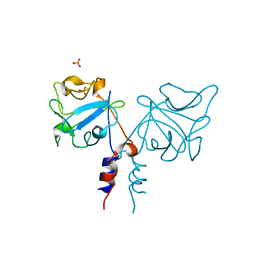 | | CRYSTAL STRUCTURE OF FIBROBLAST GROWTH FACTOR 9 (FGF9) | | Descriptor: | GLIA-ACTIVATING FACTOR, PHOSPHATE ION | | Authors: | Plotnikov, A.N, Eliseenkova, A.V, Ibrahimi, O.A, Lemmon, M.A, Mohammadi, M. | | Deposit date: | 2001-04-19 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of fibroblast growth factor 9 reveals regions implicated in dimerization and autoinhibition.
J.Biol.Chem., 276, 2001
|
|
5UTK
 
 | |
5WB7
 
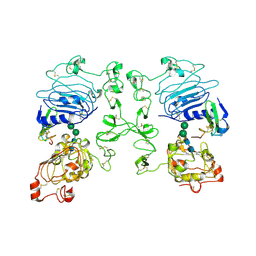 | | Crystal structure of the epidermal growth factor receptor extracellular region in complex with epiregulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Freed, D.M, Bessman, N.J, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.941 Å) | | Cite: | EGFR Ligands Differentially Stabilize Receptor Dimers to Specify Signaling Kinetics.
Cell, 171, 2017
|
|
5WB8
 
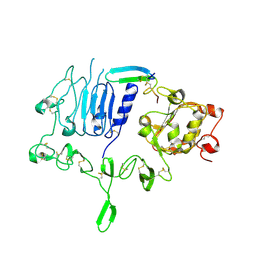 | | Crystal structure of the epidermal growth factor receptor extracellular region in complex with epigen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, Epigen, ... | | Authors: | Bessman, N.J, Freed, D.M, Moore, J.O, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | EGFR Ligands Differentially Stabilize Receptor Dimers to Specify Signaling Kinetics.
Cell, 171, 2017
|
|
7LFS
 
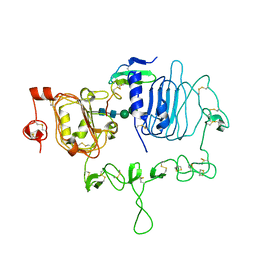 | | Crystal structure of the epidermal growth factor receptor extracellular region with A265V mutation in complex with epiregulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 4 of Epidermal growth factor receptor, ... | | Authors: | Hu, C, Leche II, C.A, Stayrook, S.E, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2021-01-18 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias.
Nature, 602, 2022
|
|
7LEN
 
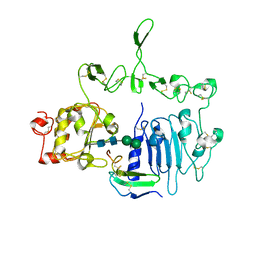 | | Crystal structure of the epidermal growth factor receptor extracellular region with R84K mutation in complex with epiregulin crystallized with trehalose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, C, Leche II, C.A, Stayrook, S.E, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2021-01-14 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias.
Nature, 602, 2022
|
|
7LFR
 
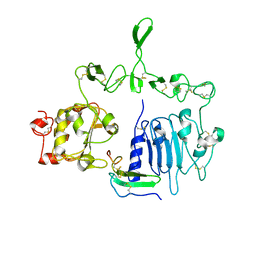 | | Crystal structure of the epidermal growth factor receptor extracellular region with R84K mutation in complex with epiregulin crystallized with spermine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, Proepiregulin, ... | | Authors: | Hu, C, Leche II, C.A, Stayrook, S.E, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2021-01-18 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias.
Nature, 602, 2022
|
|
7ME4
 
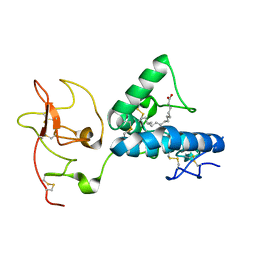 | | Structure of the extracellular WNT-binding module in Drosophila Ror2/Nrk | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITOLEIC ACID, Tyrosine-protein kinase transmembrane receptor Ror2 | | Authors: | Mendrola, J.M, Shi, F, Perry, K, Stayrook, S.E, Lemmon, M.A. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ROR and RYK extracellular region structures suggest that receptor tyrosine kinases have distinct WNT-recognition modes.
Cell Rep, 37, 2021
|
|
7ME5
 
 | | Structure of the extracellular WNT-binding module in Drl-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tyrosine-protein kinase transmembrane receptor DRL-2 | | Authors: | Shi, F, Mendrola, J.M, Perry, K, Stayrook, S.E, Lemmon, M.A. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ROR and RYK extracellular region structures suggest that receptor tyrosine kinases have distinct WNT-recognition modes.
Cell Rep, 37, 2021
|
|
4HJO
 
 | |
3GOP
 
 | |
3I2T
 
 | | Crystal structure of the unliganded Drosophila Epidermal Growth Factor Receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Alvarado, D, Klein, D.E, Lemmon, M.A. | | Deposit date: | 2009-06-29 | | Release date: | 2009-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ErbB2 resembles an autoinhibited invertebrate epidermal growth factor receptor.
Nature, 461, 2009
|
|
4GT4
 
 | |
