8TZ8
 
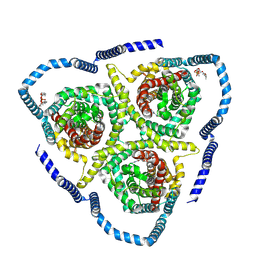 | |
5U9W
 
 | | Structure of CNTnw N149L in the intermediate 3 state | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]methyl}-2-octyldecyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, Nucleoside permease | | Authors: | Hirschi, M, Johnson, Z.L, Lee, S.-Y. | | Deposit date: | 2016-12-18 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.555 Å) | | Cite: | Visualizing multistep elevator-like transitions of a nucleoside transporter.
Nature, 545, 2017
|
|
7TX7
 
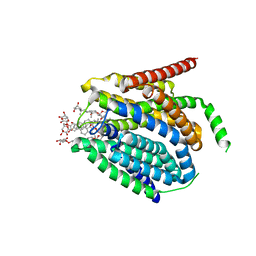 | |
7TX6
 
 | | Cryo-EM structure of the human reduced folate carrier in complex with methotrexate | | Descriptor: | Digitonin, METHOTREXATE, Reduced folate transporter,Soluble cytochrome b562 | | Authors: | Wright, N.J, Fedor, J.G, Lee, S.-Y. | | Deposit date: | 2022-02-07 | | Release date: | 2022-09-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Methotrexate recognition by the human reduced folate carrier SLC19A1.
Nature, 609, 2022
|
|
1J56
 
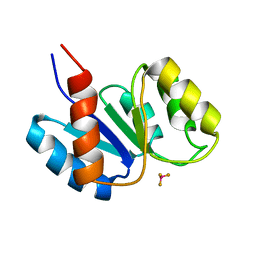 | | MINIMIZED AVERAGE STRUCTURE OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN: MODEL STRUCTURE INCORPORATING ACTIVE SITE CONTACTS | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
8ET7
 
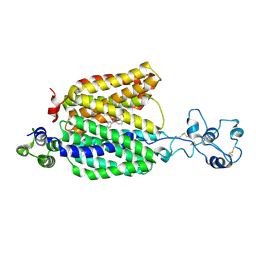 | | Cryo-EM structure of the organic cation transporter 1 in complex with diphenhydramine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-[2-(BENZHYDRYLOXY)ETHYL]-N,N-DIMETHYLAMINE, OCT1 | | Authors: | Suo, Y, Wright, N.J, Lee, S.-Y. | | Deposit date: | 2022-10-16 | | Release date: | 2023-05-31 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Molecular basis of polyspecific drug and xenobiotic recognition by OCT1 and OCT2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8ET8
 
 | | Cryo-EM structure of the organic cation transporter 1 in complex with verapamil | | Descriptor: | (2S)-2-(3,4-dimethoxyphenyl)-5-{[2-(3,4-dimethoxyphenyl)ethyl](methyl)amino}-2-(propan-2-yl)pentanenitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, OCT1 | | Authors: | Suo, Y, Wright, N.J, Lee, S.-Y. | | Deposit date: | 2022-10-16 | | Release date: | 2023-05-31 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Molecular basis of polyspecific drug and xenobiotic recognition by OCT1 and OCT2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8ET9
 
 | |
8ET6
 
 | |
6NC6
 
 | | Lipid II flippase MurJ, inward closed conformation | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Lipid II flippase MurJ, ... | | Authors: | Kuk, A.C.Y, Lee, S.-Y. | | Deposit date: | 2018-12-10 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Visualizing conformation transitions of the Lipid II flippase MurJ.
Nat Commun, 10, 2019
|
|
6NC7
 
 | | Lipid II flippase MurJ, inward open conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lipid II flippase MurJ, SULFATE ION | | Authors: | Kuk, A.C.Y, Lee, S.-Y. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Visualizing conformation transitions of the Lipid II flippase MurJ.
Nat Commun, 10, 2019
|
|
8DEP
 
 | | Cryo-EM structure of the human reduced folate carrier, apo condition | | Descriptor: | Digitonin, Reduced folate transporter,Soluble cytochrome b562 | | Authors: | Wright, N.J, Fedor, J.G, Lee, S.-Y. | | Deposit date: | 2022-06-21 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Methotrexate recognition by the human reduced folate carrier SLC19A1.
Nature, 609, 2022
|
|
6NC8
 
 | | Lipid II flippase MurJ, inward occluded conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lipid II flippase MurJ, PENTAETHYLENE GLYCOL, ... | | Authors: | Kuk, A.C.Y, Lee, S.-Y. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Visualizing conformation transitions of the Lipid II flippase MurJ.
Nat Commun, 10, 2019
|
|
6NC9
 
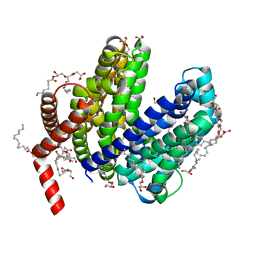 | | Lipid II flippase MurJ, outward-facing conformation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Lipid II flippase MurJ, ... | | Authors: | Kuk, A.C.Y, Lee, S.-Y. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Visualizing conformation transitions of the Lipid II flippase MurJ.
Nat Commun, 10, 2019
|
|
6NR4
 
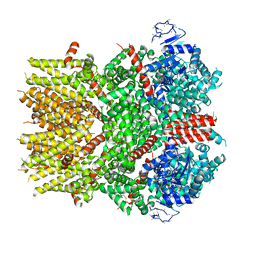 | | Cryo-EM structure of the TRPM8 ion channel with low occupancy icilin, PI(4,5)P2, and calcium | | Descriptor: | Transient receptor potential cation channel subfamily M member 8 | | Authors: | Yin, Y, Le, S.C, Hsu, A.L, Borgnia, M.J, Yang, H, Lee, S.-Y. | | Deposit date: | 2019-01-22 | | Release date: | 2019-02-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel.
Science, 363, 2019
|
|
6NR2
 
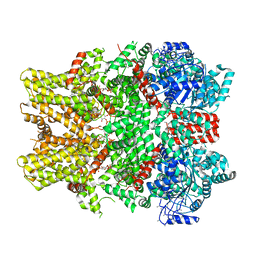 | | Cryo-EM structure of the TRPM8 ion channel in complex with the menthol analog WS-12 and PI(4,5)P2 | | Descriptor: | (1R,2S,5R)-N-(4-methoxyphenyl)-5-methyl-2-(propan-2-yl)cyclohexane-1-carboxamide, (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl icosa-5,8,11,14-tetraenoate, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Yin, Y, Le, S.C, Hsu, A.L, Borgnia, M.J, Yang, H, Lee, S.-Y. | | Deposit date: | 2019-01-22 | | Release date: | 2019-02-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel.
Science, 363, 2019
|
|
8E4O
 
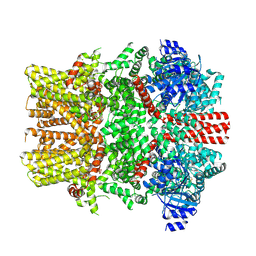 | | The closed C1-state mouse TRPM8 structure in complex with putative PI(4,5)P2 | | Descriptor: | Transient receptor potential cation channel subfamily M member 8, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
8E4N
 
 | | The closed C1-state mouse TRPM8 structure in complex with PI(4,5)P2 | | Descriptor: | Transient receptor potential cation channel subfamily M member 8, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
8E4L
 
 | | The open state mouse TRPM8 structure in complex with the cooling agonist C3, AITC, and PI(4,5)P2 | | Descriptor: | 3-isothiocyanatoprop-1-ene, CALCIUM ION, Transient receptor potential cation channel subfamily M member 8, ... | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
8E4Q
 
 | | The closed C0-state flycatcher TRPM8 structure in complex with PI(4,5)P2 | | Descriptor: | Transient receptor potential cation channel subfamily M member 8, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
8E4P
 
 | | Mouse TRPM8 structure determined in the ligand- and PI(4,5)P2-free condition, Class I , C0 state | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
8E4M
 
 | | The intermediate C2-state mouse TRPM8 structure in complex with the cooling agonist C3 and PI(4,5)P2 | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily M member 8, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Yin, Y, Zhang, F, Feng, S, Butay, K.J, Borgnia, M.J, Im, W, Lee, S.-Y. | | Deposit date: | 2022-08-18 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Activation mechanism of the mouse cold-sensing TRPM8 channel by cooling agonist and PIP 2.
Science, 378, 2022
|
|
6OO5
 
 | | Cryo-EM structure of the C2-symmetric TRPV2/RTx complex in amphipol resolved to 4.2 A | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
6OO3
 
 | | Cryo-EM structure of the C4-symmetric TRPV2/RTx complex in amphipol resolved to 2.9 A | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
6OO4
 
 | | Cryo-EM structure of the C2-symmetric TRPV2/RTx complex in amphipol resolved to 3.3 A | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
