4LO7
 
 | | HA70(D3)-HA17-HA33 | | Descriptor: | HA-17, HA-33, HA-70 | | Authors: | Lee, K, Gu, S, Jin, L, Le, T.T, Cheng, L.W, Strotmeier, J, Kruel, A.M, Yao, G, Perry, K, Rummel, A, Jin, R. | | Deposit date: | 2013-07-12 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.73 Å) | | Cite: | Structure of a Bimodular Botulinum Neurotoxin Complex Provides Insights into Its Oral Toxicity.
Plos Pathog., 9, 2013
|
|
4LO8
 
 | | HA70(D3)-HA17 | | Descriptor: | HA-17, HA-70 | | Authors: | Lee, K, Gu, S, Jin, L, Le, T.T, Cheng, L.W, Strotmeier, J, Kruel, A.M, Yao, G, Perry, K, Rummel, A, Jin, R. | | Deposit date: | 2013-07-12 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Bimodular Botulinum Neurotoxin Complex Provides Insights into Its Oral Toxicity.
Plos Pathog., 9, 2013
|
|
4OD0
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea | | Descriptor: | 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morisseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
4OCZ
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-isobutyrylpiperidin-4-yl)-3-(4-(trifluoromethyl)phenyl)urea | | Descriptor: | 1-[1-(2-methylpropanoyl)piperidin-4-yl]-3-[4-(trifluoromethyl)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morriseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
2Z3X
 
 | |
2G04
 
 | | Crystal structure of fatty acid-CoA racemase from Mycobacterium tuberculosis H37Rv | | Descriptor: | PROBABLE FATTY-ACID-CoA RACEMASE FAR | | Authors: | Lee, K.S, Park, S.M, Rhee, K.H, Bang, W.G, Hwang, K.Y, Chi, Y.M. | | Deposit date: | 2006-02-11 | | Release date: | 2007-01-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of fatty acid-CoA racemase from Mycobacterium tuberculosis H37Rv
Proteins, 64, 2006
|
|
2LXR
 
 | | Solution structure of HP1264 from Helicobacter pylori | | Descriptor: | NADH dehydrogenase I subunit E | | Authors: | Lee, K. | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HP1264, Complex I subunit E from Helicobacter pylori, reveals a novel fold for FMN binding protein
To be Published
|
|
7RSE
 
 | | NMR-driven structure of the KRAS4B-G12D "alpha-beta" dimer on a lipid bilayer nanodisc | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
7RSC
 
 | | NMR-driven structure of the KRAS4B-G12D "alpha-alpha" dimer on a lipid bilayer nanodisc | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | Authors: | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | Deposit date: | 2021-08-11 | | Release date: | 2021-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
7L7J
 
 | | Cryo-EM structure of Hsp90:p23 closed-state complex | | Descriptor: | Heat shock protein HSP 90-alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Prostaglandin E synthase 3 | | Authors: | Lee, K, Thwin, A.C, Tse, E, Gates, S.N, Southworth, D.R. | | Deposit date: | 2020-12-28 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The structure of an Hsp90-immunophilin complex reveals cochaperone recognition of the client maturation state.
Mol.Cell, 81, 2021
|
|
7L7I
 
 | | Cryo-EM structure of Hsp90:FKBP51:p23 closed-state complex | | Descriptor: | Heat shock protein HSP 90-alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Peptidyl-prolyl cis-trans isomerase FKBP5, ... | | Authors: | Lee, K, Thwin, A.C, Tse, E, Gates, S.N, Southworth, D.R. | | Deposit date: | 2020-12-28 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of an Hsp90-immunophilin complex reveals cochaperone recognition of the client maturation state.
Mol.Cell, 81, 2021
|
|
8CRC
 
 | | Structure of human Plk1 PBD in complex with Allopole-A | | Descriptor: | 7-chloro-4-(cyclopropylmethyl)-1-thioxo-2,4-dihydrothieno[2,3-e][1,2,4]triazolo[4,3-a]pyrimidin-5(1H)-one, GLYCEROL, Serine/threonine-protein kinase PLK1 | | Authors: | Kirsch, K, Park, J.E, Lee, K.S. | | Deposit date: | 2023-03-08 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Specific inhibition of an anticancer target, polo-like kinase 1, by allosterically dismantling its mechanism of substrate recognition.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1QZ0
 
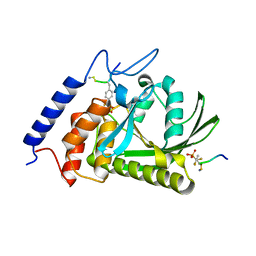 | | Crystal Structure of the Yersinia Pestis Phosphatase YopH in Complex with a Phosphotyrosyl Mimetic-Containing Hexapeptide | | Descriptor: | ASP-ALA-ASP-GLU-FTY-LEU-NH2, Protein-tyrosine phosphatase yopH | | Authors: | Phan, J, Lee, K, Cherry, S, Tropea, J.E, Burke Jr, T.R, Waugh, D.S. | | Deposit date: | 2003-09-15 | | Release date: | 2003-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-Resolution Structure of the Yersinia pestis Protein Tyrosine Phosphatase YopH in Complex with a Phosphotyrosyl Mimetic-Containing Hexapeptide
Biochemistry, 42, 2003
|
|
2AP9
 
 | | Crystal structure of acetylglutamate kinase from Mycobacterium tuberculosis CDC1551 | | Descriptor: | MAGNESIUM ION, NICKEL (II) ION, acetylglutamate kinase | | Authors: | Rajashankar, K.R, Kniewel, R, Lee, K, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-08-15 | | Release date: | 2005-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of acetylglutamate kinase from Mycobacterium tuberculosis CDC1551
To be Published
|
|
4N7Z
 
 | | Crystal structure of human Plk4 cryptic polo box (CPB) in complex with a Cep192 N-terminal fragment | | Descriptor: | Centrosomal protein of 192 kDa, Serine/threonine-protein kinase PLK4 | | Authors: | Park, S.-Y, Park, J.-E, Tian, L, Kim, T.-S, Yang, W, Lee, K.S. | | Deposit date: | 2013-10-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5X03
 
 | |
8JTD
 
 | | BJOX2000.664 trimer in complex with Fab fragment of broadly neutralizing HIV antibody PGT145 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PGT145 antibody fragment, ... | | Authors: | Chatterjee, A, Chen, C, Lee, K, Mangala Prasad, V. | | Deposit date: | 2023-06-21 | | Release date: | 2023-10-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | An HIV-1 broadly neutralizing antibody overcomes structural and dynamic variation through highly focused epitope targeting.
Npj Viruses, 1, 2023
|
|
8JTM
 
 | | CNE55.664 trimer in complex with broadly neutralizing HIV antibody PGT145 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PGT145 antibody fragment, ... | | Authors: | Chatterjee, A, Chen, C, Lee, K, Mangala Prasad, V. | | Deposit date: | 2023-06-22 | | Release date: | 2023-10-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.14 Å) | | Cite: | An HIV-1 broadly neutralizing antibody overcomes structural and dynamic variation through highly focused epitope targeting.
Npj Viruses, 1, 2023
|
|
4WHK
 
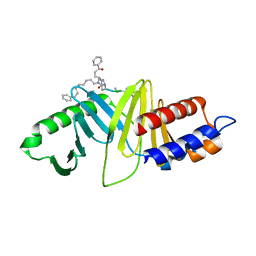 | | A New Class of Peptidomimetics Targeting the Polo-box Domain of Polo-like kinase 1 | | Descriptor: | C6H5(CH2)8-DERIVATIZED PEPTIDE INHIBITOR, Serine/threonine-protein kinase PLK1 | | Authors: | Bang, J.K, Han, Y.H, Ahn, M.J, Lee, K.S. | | Deposit date: | 2014-09-23 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A new class of peptidomimetics targeting the polo-box domain of polo-like kinase 1.
J.Med.Chem., 58, 2015
|
|
4WHL
 
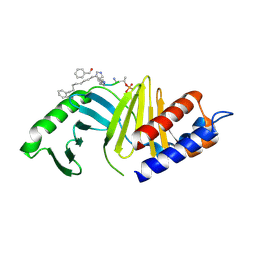 | | A New Class of Peptidomimetics Targeting the Polo-box Domain of Polo-like kinase 1 | | Descriptor: | C6H5(CH2)8-DERIVATIZED PEPTIDE INHIBITOR, Serine/threonine-protein kinase PLK1 | | Authors: | Bang, J.K, Han, Y.H, Ahn, M.J, Lee, K.S. | | Deposit date: | 2014-09-23 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A new class of peptidomimetics targeting the polo-box domain of polo-like kinase 1.
J.Med.Chem., 58, 2015
|
|
4NQY
 
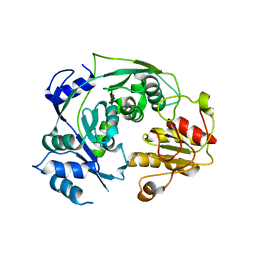 | | The reduced form of MJ0499 | | Descriptor: | Isopropylmalate/citramalate isomerase large subunit, ZINC ION | | Authors: | Hwang, K.Y, Lee, E.H, Lee, K. | | Deposit date: | 2013-11-26 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural characterization and comparison of the large subunits of IPM isomerase and homoaconitase from Methanococcus jannaschii
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6CSV
 
 | | The structure of the Cep63-Cep152 heterotetrameric complex | | Descriptor: | Centrosomal protein of 63 kDa,Centrosomal protein of 152 kDa | | Authors: | Lee, E, Chen, Y, Zhang, L, Kim, T.S, Ahn, J.I, Park, J.E, Lee, K.S. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular architecture of a cylindrical self-assembly at human centrosomes.
Nat Commun, 10, 2019
|
|
6CSU
 
 | | The structure of the Cep63-Cep152 heterotetrameric complex | | Descriptor: | Centrosomal protein of 152 kDa, Centrosomal protein of 63 kDa | | Authors: | Lee, E, Chen, Y, Zhang, L, Kim, T.S, Ahn, J.I, Park, J.E, Lee, K.S. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular architecture of a cylindrical self-assembly at human centrosomes.
Nat Commun, 10, 2019
|
|
9FLU
 
 | | CryoEM structure of the fragment-1 (3402-3733) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
9FLX
 
 | | CryoEM structure of the fragment-4 (4074-4421) in the grappling hook protein A (GhpA) in the bacterium Aureispira sp. CCB-QB1 | | Descriptor: | The grappling hook protein A in the bacterium Aureispira sp. CCB-QB1 | | Authors: | Lien, Y.-W, Amendola, D, Lee, K.S, Bartlau, N, Xu, J, Furusawa, G, Polz, M.F, Stocker, R, Weiss, G.L, Pilhofer, M. | | Deposit date: | 2024-06-05 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of bacterial predation via ixotrophy.
Science, 386, 2024
|
|
