8PJ8
 
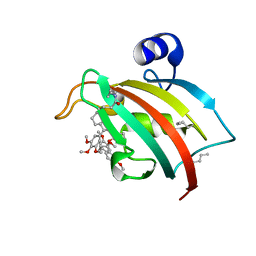 | | FKBP51FK1 F67E/K60Orn (i, i+7) in complex with SAFit1 | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Charalampidou, A, Hausch, F. | | Deposit date: | 2023-06-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Automated Flow Peptide Synthesis Enables Engineering of Proteins with Stabilized Transient Binding Pockets.
Acs Cent.Sci., 10, 2024
|
|
8PJA
 
 | | FKBP51FK1 F67E/K58 (i, i+9) in complex with SAFit1 | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Charalampidou, A, Hausch, F. | | Deposit date: | 2023-06-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Automated Flow Peptide Synthesis Enables Engineering of Proteins with Stabilized Transient Binding Pockets.
Acs Cent.Sci., 10, 2024
|
|
1IF2
 
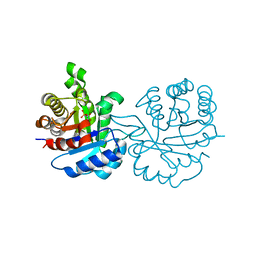 | | X-RAY STRUCTURE OF LEISHMANIA MEXICANA TRIOSEPHOSPHATE ISOMERASE COMPLEXED WITH IPP | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE, [2(FORMYL-HYDROXY-AMINO)-ETHYL]-PHOSPHONIC ACID | | Authors: | Kursula, I, Partanen, S, Lambeir, A.-M, Antonov, D.M, Augustyns, K, Wierenga, R.K. | | Deposit date: | 2001-04-12 | | Release date: | 2001-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural determinants for ligand binding and catalysis of triosephosphate isomerase.
Eur.J.Biochem., 268, 2001
|
|
3O6O
 
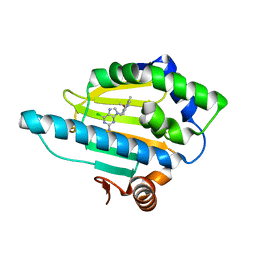 | | Crystal Structure of the N-terminal domain of an HSP90 from Trypanosoma Brucei, Tb10.26.1080 in the presence of an the inhibitor BIIB021 | | Descriptor: | 6-chloro-9-[(4-methoxy-3,5-dimethylpyridin-2-yl)methyl]-9H-purin-2-amine, Heat shock protein 83, PENTAETHYLENE GLYCOL | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, Weadge, J, Li, Y, Kozieradzki, I, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Wyatt, P, Fairlamb, A.H, Ferguson, M.A.J, Thompson, S, MacKenzie, C, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the Trypanosoma brucei Hsp83 potential as a target for structure guided drug design.
PLoS Negl Trop Dis, 7, 2013
|
|
2WOI
 
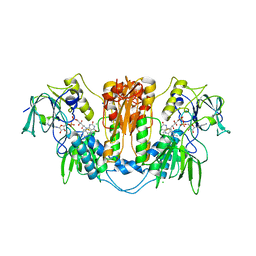 | | Trypanothione reductase from Trypanosoma brucei | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Alphey, M.S, Fairlamb, A.H. | | Deposit date: | 2009-07-24 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
3OMU
 
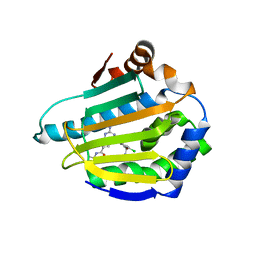 | | Crystal Structure of the N-terminal domain of an HSP90 from Trypanosoma Brucei, Tb10.26.1080 in the presence of a thienopyrimidine derivative | | Descriptor: | 2-amino-4-{2,4-dichloro-5-[2-(diethylamino)ethoxy]phenyl}-N-ethylthieno[2,3-d]pyrimidine-6-carboxamide, Heat shock protein 83 | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, Weadge, J, Cossar, D, Li, Y, Kozieradzki, I, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Wyatt, P.G, Fairlamb, A.H, MacKenzie, C, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-27 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Exploring the Trypanosoma brucei Hsp83 potential as a target for structure guided drug design.
PLoS Negl Trop Dis, 7, 2013
|
|
2WP6
 
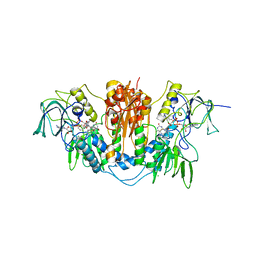 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00071494) | | Descriptor: | (4S)-3-BENZYL-6-CHLORO-2-METHYL-4-PHENYL-3,4-DIHYDROQUINAZOLINE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-03 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
2WPF
 
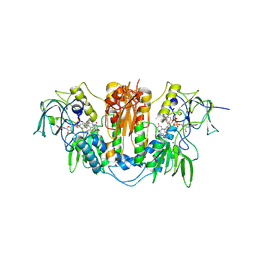 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00085762) | | Descriptor: | 3-[(4S)-6-CHLORO-2-METHYL-4-(4-METHYLPHENYL)QUINAZOLIN-3(4H)-YL]-N,N-DIMETHYLPROPAN-1-AMINE, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-06 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
2WSA
 
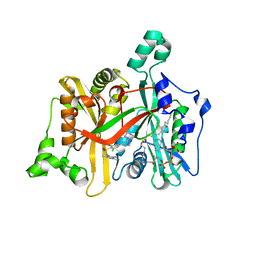 | | Crystal Structure of Leishmania major N-myristoyltransferase (NMT) with bound myristoyl-CoA and a pyrazole sulphonamide ligand (DDD85646) | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Brand, S, Fairlamb, A.H, Ferguson, M.A.J, Frearson, J.A, Wyatt, P.G, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-04 | | Release date: | 2010-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | N-Myristoyltransferase Inhibitors as New Leads to Treat Sleeping Sickness.
Nature, 464, 2010
|
|
3OPD
 
 | | Crystal Structure of the N-terminal domain of an HSP90 from Trypanosoma Brucei, Tb10.26.1080 in the presence of a benzamide derivative | | Descriptor: | 4-[6,6-dimethyl-4-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl]-2-[(cis-4-hydroxycyclohexyl)amino]benzamide, Heat shock protein 83 | | Authors: | Pizarro, J.C, Wernimont, A.K, Hutchinson, A, Sullivan, H, Chamberlain, K, Weadge, J, Cossar, D, Li, Y, Kozieradzki, I, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Wyatt, P.G, Fairlamb, A.H, MacKenzie, C, Ferguson, M.A.J, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-31 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exploring the Trypanosoma brucei Hsp83 potential as a target for structure guided drug design.
PLoS Negl Trop Dis, 7, 2013
|
|
2WPC
 
 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00073357) | | Descriptor: | (4S)-6-CHLORO-3-{2-[4-(FURAN-2-YLCARBONYL)PIPERAZIN-1-YL]ETHYL}-2-METHYL-4-PHENYL-3,4-DIHYDROQUINAZOLINE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-05 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
2WOV
 
 | | Trypanosoma brucei trypanothione reductase with bound NADP. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, ... | | Authors: | Alphey, M.S, Fairlamb, A.H. | | Deposit date: | 2009-07-30 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
2WPE
 
 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00073359) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-06 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
2WOW
 
 | | Trypanosoma brucei trypanothione reductase with NADP and trypanothione bound | | Descriptor: | BIS(GAMMA-GLUTAMYL-CYSTEINYL-GLYCINYL)SPERMIDINE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alphey, M.S, Fairlamb, A.H. | | Deposit date: | 2009-07-30 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
2WP5
 
 | | Trypanosoma brucei trypanothione reductase in complex with 3,4- dihydroquinazoline inhibitor (DDD00065414) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Alphey, M.S, Patterson, S, Fairlamb, A.H. | | Deposit date: | 2009-08-03 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dihydroquinazolines as a Novel Class of Trypanosoma Brucei Trypanothione Reductase Inhibitors: Discovery, Synthesis, and Characterization of Their Binding Mode by Protein Crystallography.
J.Med.Chem., 54, 2011
|
|
2VOB
 
 | | TRYPANOTHIONE SYNTHETASE | | Descriptor: | CHLORIDE ION, TRYPANOTHIONE SYNTHETASE | | Authors: | Fyfe, P.K, Oza, S.L, Fairlamb, A.H, Hunter, W.N. | | Deposit date: | 2008-02-13 | | Release date: | 2008-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Leishmania Trypanothione Synthetase-Amidase Structure Reveals a Basis for Regulation of Conflicting Synthetic and Hydrolytic Activities.
J.Biol.Chem., 283, 2008
|
|
3PEH
 
 | | Crystal Structure of the N-terminal domain of an HSP90 from Plasmodium Falciparum, PFL1070c in the presence of a thienopyrimidine derivative | | Descriptor: | 2-amino-4-{2,4-dichloro-5-[2-(diethylamino)ethoxy]phenyl}-N-ethylthieno[2,3-d]pyrimidine-6-carboxamide, Endoplasmin homolog, SULFATE ION | | Authors: | Wernimont, A.K, Tempel, W, Hutchinson, A, Weadge, J, Cossar, D, MacKenzie, F, Vedadi, M, Senisterra, G, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Wyatt, P.G, Fairlamb, A.H, MacKenzie, C, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-26 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of the N-terminal domain of an HSP90 from Plasmodium Falciparum, PFL1070c in the presence of a thienopyrimidine derivative
To be Published
|
|
3OGZ
 
 | | Protein structure of USP from L. major in Apo-form | | Descriptor: | GLYCEROL, UDP-sugar pyrophosphorylase | | Authors: | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F, Ficner, R. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
2XIM
 
 | | ARGININE RESIDUES AS STABILIZING ELEMENTS IN PROTEINS | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION, Xylitol | | Authors: | Mrabet, N.T, Van Denbroek, A, Van Den Brande, I, Stanssens, P, Laroche, Y, Lambeir, A.-M, Matthyssens, G, Jenkins, J, Chiadmi, M, Vantilbeurgh, H, Rey, F, Janin, J, Quax, W.J, Lasters, I, Demaeyer, M, Wodak, S.J. | | Deposit date: | 1991-05-29 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Arginine residues as stabilizing elements in proteins.
Biochemistry, 31, 1992
|
|
3PP4
 
 | |
3PP3
 
 | |
3Q5K
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of an inhibitor | | Descriptor: | 4-[6,6-dimethyl-4-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl]-2-{[2-(methylsulfanyl)ethyl]amino}benzamide, Heat shock protein 83-1 | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-28 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of an inhibitor
To be Published
|
|
3Q5L
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K 213 in the presence of 17-AEP-geldanamycin | | Descriptor: | (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-13-hydroxy-8,14-dimethoxy-4,10,12,16-tetramethyl-3,20,22-trioxo-19-{[2-(pyrrolidin-1-yl)ethyl]amino}-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate, Heat shock protein 83-1 | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-28 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K 213 in the presence of 17-AEP-geldanamycin.
To be Published
|
|
3Q5J
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of 17-DMAP-geldanamycin | | Descriptor: | (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-19-{[3-(dimethylamino)propyl]amino}-13-hydroxy-8,14-dimethoxy-4,10,12,16-tetramethyl-3,20,22-trioxo-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate, Heat shock protein 83-1 | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-28 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of 17-DMAP-geldanamycin.
To be Published
|
|
3OH2
 
 | | Protein structure of USP from L. major bound to URIDINE-5'-DIPHOSPHATE-GALACTOSE | | Descriptor: | GALACTOSE-URIDINE-5'-DIPHOSPHATE, GLYCEROL, UDP-sugar pyrophosphorylase | | Authors: | Dickmanns, A, Damerow, S, Neumann, P, Schulz, E.-C, Lamerz, A, Routier, F, Ficner, R. | | Deposit date: | 2010-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural basis for the broad substrate range of the UDP-sugar pyrophosphorylase from Leishmania major.
J.Mol.Biol., 405, 2011
|
|
