8UGY
 
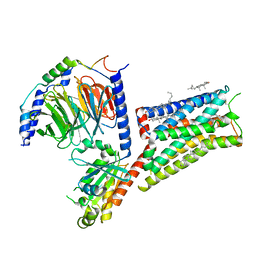 | | Serotonin 1E receptor (5-HT1eR)-Gi1 Complex bound with Mianserin | | Descriptor: | 5-hydroxytryptamine receptor 1E, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zilberg, G, Warren, A.L, Wacker, D. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural insights into the unexpected agonism of tetracyclic antidepressants at serotonin receptors 5-HT 1e R and 5-HT 1F R.
Sci Adv, 10, 2024
|
|
5TPJ
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | denovo NTF2 | | Authors: | Basanta, B, Oberdorfer, G, Marcos, E, Chidyausiku, T.M, Sankaran, B, Baker, D. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
8VC8
 
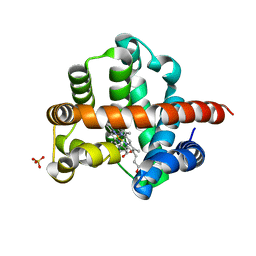 | |
1MHX
 
 | | Crystal Structures of the redesigned protein G variant NuG1 | | Descriptor: | immunoglobulin-binding protein G | | Authors: | Nauli, S, Kuhlman, B, Le Trong, I, Stenkamp, R.E, Teller, D.C, Baker, D. | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and increased stabilization of the protein G variants with switched folding pathways NuG1 and NuG2
Protein Sci., 11, 2002
|
|
5AD2
 
 | | Bivalent binding to BET bromodomains | | Descriptor: | (3R)-4-(2-{4-[1-(3-chloro[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidinyl]phenoxy}ethyl)-1,3-dimethyl-2-piperazinone, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
5TPH
 
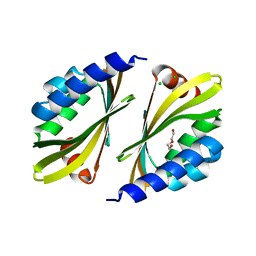 | | Crystal structure of a de novo designed protein homodimer with curved beta-sheet | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, de novo NTF2 homodimer | | Authors: | Basanta, B, Marcos, E, Oberdorfer, G, Chidyausiku, T.M, Sankaran, B, Baker, D. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5TS4
 
 | | Crystal structure of a de novo designed protein with curved beta-sheet | | Descriptor: | DI(HYDROXYETHYL)ETHER, denovo NTF2 | | Authors: | Basanta, B, Oberdorfer, G, Chidyausiku, T.M, Marcos, E, Pereira, J.H, Sankaran, B, Zwart, P.H, Baker, D. | | Deposit date: | 2016-10-27 | | Release date: | 2017-01-25 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5AD3
 
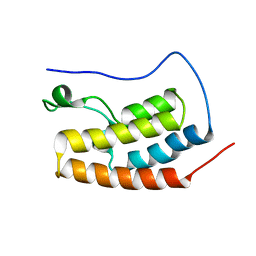 | | Bivalent binding to BET bromodomains | | Descriptor: | 3-methoxy-N-[2-[4-[1-(3-methoxy-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidyl]phenoxy]ethyl]-N-methyl-[1,2,4]triazolo[4,3-b]pyridazin-6-amine, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
5UYO
 
 | | Solution NMR structure of the de novo mini protein HEEH_rd4_0097 | | Descriptor: | HEEH_rd4_0097 | | Authors: | Lemak, A, Rocklin, G.J, Houliston, S, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-02-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
5UP5
 
 | | Solution structure of the de novo mini protein EHEE_rd1_0284 | | Descriptor: | EHEE_rd1_0284 | | Authors: | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
5W9F
 
 | | Solution structure of the de novo mini protein gHEEE_02 | | Descriptor: | De novo mini protein gHEEE_02 | | Authors: | Pulavarti, S.V.S.R.K, Shaw, E.A, Bahl, C.D, Garry, B.W, Baker, D, Szyperski, T. | | Deposit date: | 2017-06-23 | | Release date: | 2018-07-11 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Cytosolic expression, solution structures, and molecular dynamics simulation of genetically encodable disulfide-rich de novo designed peptides.
Protein Sci., 27, 2018
|
|
3J03
 
 | | Lidless Mm-cpn in the closed state with ATP/AlFx | | Descriptor: | Lidless Mm-cpn | | Authors: | Zhang, J, Ma, B, DiMaio, F, Douglas, N.R, Joachimiak, L, Baker, D, Frydman, J, Levitt, M, Chiu, W. | | Deposit date: | 2011-02-10 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM structure of a group II chaperonin in the prehydrolysis ATP-bound state leading to lid closure.
Structure, 19, 2011
|
|
4LT9
 
 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR404 | | Descriptor: | Engineered Protein OR404 | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Sahdev, S, Xiao, R, Kogan, S, Maglaqui, M, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Engineered Protein OR404.
To be Published
|
|
4ESS
 
 | | Crystal Structure of E6D/L155R variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR187 | | Descriptor: | OR187 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9971 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4ETK
 
 | | Crystal Structure of E6A/L130D/A155H variant of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR186 | | Descriptor: | De novo designed serine hydrolase, SODIUM ION | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4NWQ
 
 | | Computationally Designed Two-Component Self-Assembling Tetrahedral Cage, T33-21, Crystallized in Space Group F4132 | | Descriptor: | Putative uncharacterized protein PH0671, SULFATE ION, Uncharacterized protein | | Authors: | McNamara, D.E, King, N.P, Bale, J.B, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2013-12-06 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Accurate design of co-assembling multi-component protein nanomaterials.
Nature, 510, 2014
|
|
4NE7
 
 | | Crystal Structure of engineered Kumamolisin-As from Alicyclobacillus sendaiensis, Northeast Structural Genomics Consortium (NESG) Target OR367 | | Descriptor: | Kumamolisin-As, ZINC ION | | Authors: | Guan, R, Pultz, I.S, Siegel, J.B, Seetharaman, J, Kornhaber, G, Maglaqui, M, Mao, L, Xiao, R, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-28 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR367
To be Published
|
|
4NWP
 
 | | Computationally Designed Two-Component Self-Assembling Tetrahedral Cage, T33-21, Crystallized in Space Group R32 | | Descriptor: | AMMONIUM ION, Putative uncharacterized protein, SULFATE ION, ... | | Authors: | McNamara, D.E, King, N.P, Bale, J.B, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2013-12-06 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Accurate design of co-assembling multi-component protein nanomaterials.
Nature, 510, 2014
|
|
4NWR
 
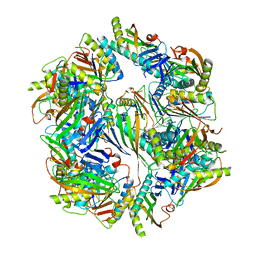 | | Computationally Designed Two-Component Self-Assembling Tetrahedral Cage T33-28 | | Descriptor: | Macrophage migration inhibitory factor-like protein, integron gene cassette protein | | Authors: | McNamara, D.E, King, N.P, Bale, J.B, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2013-12-06 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Accurate design of co-assembling multi-component protein nanomaterials.
Nature, 510, 2014
|
|
4NWO
 
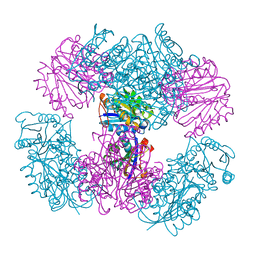 | | Computationally Designed Two-Component Self-Assembling Tetrahedral Cage T33-15 | | Descriptor: | CALCIUM ION, Chorismate mutase AroH, Molybdenum cofactor biosynthesis protein MogA | | Authors: | McNamara, D.E, King, N.P, Bale, J.B, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2013-12-06 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Accurate design of co-assembling multi-component protein nanomaterials.
Nature, 510, 2014
|
|
1MOW
 
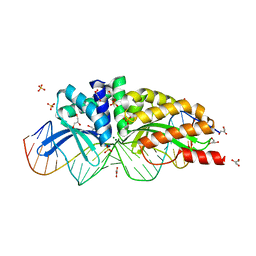 | | E-DreI | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*CP*TP*GP*TP*CP*TP*CP*AP*AP*GP*TP*TP*CP*CP*GP*GP*CP*G)-3', 5'-D(*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*GP*AP*GP*AP*CP*AP*GP*TP*TP*TP*GP*G)-3', GLYCEROL, ... | | Authors: | Chevalier, B.S, Kortemme, T, Chadsey, M.S, Baker, D, Monnat Jr, R.J, Stoddard, B.L. | | Deposit date: | 2002-09-10 | | Release date: | 2002-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design, Activity and Structure of a Highly Specific Artificial Endonuclease
Mol.Cell, 10, 2002
|
|
1MPU
 
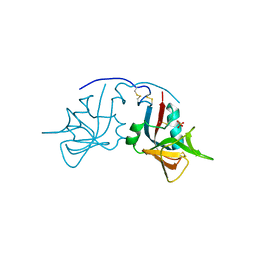 | | Crystal Structure of the free human NKG2D immunoreceptor | | Descriptor: | NKG2-D type II integral membrane protein, PHOSPHATE ION | | Authors: | McFarland, B.J, Kortemme, T, Baker, D, Strong, R.K. | | Deposit date: | 2002-09-12 | | Release date: | 2003-04-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Symmetry Recognizing Asymmetry: Analysis of the Interactions between the C-Type Lectin-like Immunoreceptor NKG2D and MHC
Class I-like Ligands
Structure, 11, 2003
|
|
7LMX
 
 | | A HIGHLY SPECIFIC INHIBITOR OF INTEGRIN ALPHA-V BETA-6 WITH A DISULFIDE | | Descriptor: | Integrin inhibitor | | Authors: | Dong, X, Bera, A.K, Roy, A, Shi, L, Springer, T.A, Baker, D. | | Deposit date: | 2021-02-06 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
7LMV
 
 | | SPECIFIC INHIBITOR OF INTEGRIN ALPHA-V BETA-6 | | Descriptor: | Integrin inhibitor | | Authors: | Dong, X, Bera, A.K, Roy, A, Shi, L, Springer, T.A, Baker, D. | | Deposit date: | 2021-02-05 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo design of highly selective miniprotein inhibitors of integrins alpha v beta 6 and alpha v beta 8.
Nat Commun, 14, 2023
|
|
2ERH
 
 | | Crystal Structure of the E7_G/Im7_G complex; a designed interface between the colicin E7 DNAse and the Im7 immunity protein | | Descriptor: | Colicin E7, Colicin E7 immunity protein | | Authors: | Joachimiak, L.A, Kortemme, T, Stoddard, B.L, Baker, D. | | Deposit date: | 2005-10-24 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational Design of a New Hydrogen Bond Network and at Least a 300-fold Specificity Switch at a Protein-Protein Interface.
J.Mol.Biol., 361, 2006
|
|
