5BOA
 
 | | Crystal Structure of the Meningitis Pathogen Streptococcus suis adhesion Fhb bound to the disaccharide receptor Gb2 | | Descriptor: | Translation initiation factor 2 (IF-2 GTPase), alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Zhang, C, Yu, Y, Yang, M, Jiang, Y. | | Deposit date: | 2015-05-27 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | Structural basis of the interaction between the meningitis pathogen Streptococcus suis adhesin Fhb and its human receptor.
Febs Lett., 590, 2016
|
|
5GOP
 
 | | Crystal structure of alkaline invertase InvA from Anabaena sp. PCC 7120 complexed with sucrose | | Descriptor: | Alkaline Invertase, beta-D-fructofuranose, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Xie, J, Cai, K, Hu, H.X, Jiang, Y.L, Yang, F, Hu, P.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Analysis of the Catalytic Mechanism and Substrate Specificity of Anabaena Alkaline Invertase InvA Reveals a Novel Glucosidase
J. Biol. Chem., 291, 2016
|
|
5HWY
 
 | | Structural mechanisms of extracellular ion exchange and induced binding-site occlusion in the sodium-calcium exchanger NCX_Mj soaked with 10 mM Na+ and zero Ca2+ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ACETATE ION, PENTADECANE, ... | | Authors: | Liao, J, Jiang, Y.X, Faraldo-Gomez, J.D. | | Deposit date: | 2016-01-29 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Mechanism of extracellular ion exchange and binding-site occlusion in a sodium/calcium exchanger
Nat.Struct.Mol.Biol., 23, 2016
|
|
3LA3
 
 | | Crystal structure of NtcA in complex with 2,2-difluoropentanedioic acid | | Descriptor: | 2,2-difluoropentanedioic acid, Global nitrogen regulator | | Authors: | Zhao, M.X, Jiang, Y.L, He, Y.X, Chen, Y.F, Teng, Y.B, Chen, Y.X, Zhang, C.C, Zhou, C.Z. | | Deposit date: | 2010-01-06 | | Release date: | 2010-07-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ERF
 
 | | Crystal structure of Gtt2 from Saccharomyces cerevisiae | | Descriptor: | Glutathione S-transferase 2 | | Authors: | Ma, X.X, Jiang, Y.L, He, Y.X, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2008-10-02 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structures of yeast glutathione-S-transferase Gtt2 reveal a new catalytic type of GST family.
Embo Rep., 10, 2009
|
|
3HIA
 
 | | Crystal structure of the choline binding domain of Spr1274 in Streptococcus pneumoniae | | Descriptor: | Choline binding protein, PHOSPHOCHOLINE, SULFATE ION | | Authors: | Zhang, Z.-Y, Li, W.-Z, Jiang, Y.-L, Bao, R, Zhou, C.-Z, Chen, Y.-X. | | Deposit date: | 2009-05-19 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of the choline binding domain of Spr1274 in Streptococcus pneumoniae
To be Published
|
|
3EYX
 
 | | Crystal structure of Carbonic Anhydrase Nce103 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Carbonic anhydrase, ... | | Authors: | Teng, Y.B, Jiang, Y.L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2008-10-22 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural insights into the substrate tunnel of Saccharomyces cerevisiae carbonic anhydrase Nce103.
Bmc Struct.Biol., 9, 2009
|
|
3LA2
 
 | | Crystal structure of NtcA in complex with 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Global nitrogen regulator | | Authors: | Zhao, M.X, Jiang, Y.L, He, Y.X, Chen, Y.F, Teng, Y.B, Chen, Y.X, Zhang, C.C, Zhou, C.Z. | | Deposit date: | 2010-01-06 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5TUA
 
 | | structure of a Na+-selective mutant of two-pore channel from Arabidopsis thaliana AtTPC1 | | Descriptor: | BARIUM ION, CALCIUM ION, SODIUM ION, ... | | Authors: | Guo, J, Zeng, W, Jiang, Y. | | Deposit date: | 2016-11-05 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Tuning the ion selectivity of two-pore channels.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4MZ4
 
 | | Discovery of an Irreversible HCV NS5B Polymerase Inhibitor | | Descriptor: | 1-[(2-chloroquinolin-3-yl)methyl]-6-fluoro-5-methyl-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxylic acid, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Zeng, Q, Anilkumar, G.N, Rosenblum, S.B, Huang, H.-C, Lesburg, C.A, Jiang, Y, Selyutin, O, Chan, T.-Y, Bennett, F, Chen, K.X, Venkatraman, S, Sannigrahi, M, Velazquez, F, Duca, J.S, Gavalas, S, Huang, Y, Pu, H, Wang, L, Pinto, P, Vibulbhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Li, C, Hesk, D, Gesell, J, Sorota, S, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2013-09-29 | | Release date: | 2013-12-11 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Discovery of an irreversible HCV NS5B polymerase inhibitor.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8YR2
 
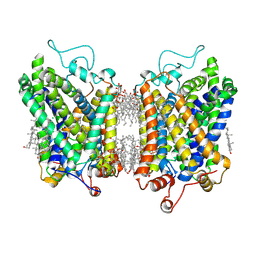 | | Structure of NET-Nisoxetine in outward-open state | | Descriptor: | CHLORIDE ION, CHOLESTEROL, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-03-20 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8X7A
 
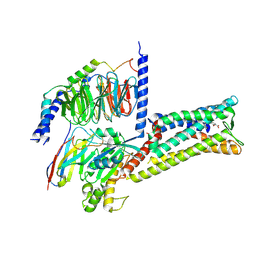 | | Treprostinil bound Prostacyclin Receptor G protein complex | | Descriptor: | 2-[[(1~{R},2~{R},3~{a}~{S},9~{a}~{S})-2-oxidanyl-1-[(3~{S})-3-oxidanyloctyl]-2,3,3~{a},4,9,9~{a}-hexahydro-1~{H}-cyclopenta[g]naphthalen-5-yl]oxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.J, Jin, S, Zhang, H, Xu, Y, Hu, W, Jiang, Y, Chen, C, Wang, D.W, Xu, H.E, Wu, C. | | Deposit date: | 2023-11-23 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Molecular recognition and activation of the prostacyclin receptor by anti-pulmonary arterial hypertension drugs.
Sci Adv, 10, 2024
|
|
8X79
 
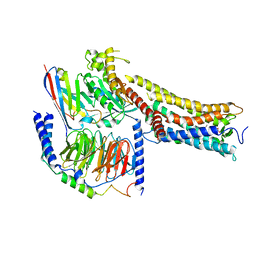 | | MRE-269 bound Prostacyclin Receptor G protein complex | | Descriptor: | 2-[4-[(5,6-diphenylpyrazin-2-yl)-propan-2-yl-amino]butoxy]ethanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.J, Jin, S, Zhang, H, Xu, Y, Hu, W, Jiang, Y, Chen, C, Wang, D.W, Xu, H.E, Wu, C. | | Deposit date: | 2023-11-23 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Molecular recognition and activation of the prostacyclin receptor by anti-pulmonary arterial hypertension drugs.
Sci Adv, 10, 2024
|
|
8SGT
 
 | |
8SGJ
 
 | |
8SGI
 
 | | Cryo-EM structure of human NCX1 in complex with SEA0400 | | Descriptor: | 2-{4-[(2,5-difluorophenyl)methoxy]phenoxy}-5-ethoxyaniline, CALCIUM ION, Fab heavy chain, ... | | Authors: | Xue, J, Jiang, Y. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanisms of human cardiac sodium calcium exchanger NCX1
To Be Published
|
|
3ERG
 
 | | Crystal structure of Gtt2 from Saccharomyces cerevisiae in complex with glutathione sulfnate | | Descriptor: | GLUTATHIONE SULFONIC ACID, Glutathione S-transferase 2 | | Authors: | Ma, X.X, Jiang, Y.L, He, Y.X, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2008-10-02 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of yeast glutathione-S-transferase Gtt2 reveal a new catalytic type of GST family.
Embo Rep., 10, 2009
|
|
3NAF
 
 | | Structure of the Intracellular Gating Ring from the Human High-conductance Ca2+ gated K+ Channel (BK Channel) | | Descriptor: | Calcium-activated potassium channel subunit alpha-1,Calcium-activated potassium channel subunit alpha-1,Calcium-activated potassium channel subunit alpha-1 | | Authors: | Wu, Y, Yang, Y, Ye, S, Jiang, Y. | | Deposit date: | 2010-06-01 | | Release date: | 2010-06-30 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the gating ring from the human large-conductance Ca(2+)-gated K(+) channel.
Nature, 466, 2010
|
|
2AEF
 
 | | Crystal Structures of the MthK RCK Domain in Ca2+ bound form | | Descriptor: | CALCIUM ION, Calcium-gated potassium channel mthK | | Authors: | Dong, J, Shi, N, Berke, I, Chen, L, Jiang, Y. | | Deposit date: | 2005-07-22 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the MthK RCK Domain and the Effect of Ca2+ on Gating Ring Stability
J.Biol.Chem., 280, 2005
|
|
8Y92
 
 | | structure of NET-Atomoxetine in outward-open state | | Descriptor: | (3R)-N-methyl-3-(2-methylphenoxy)-3-phenyl-propan-1-amine, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8Y91
 
 | | Structure of NET-nomifensine in outward-open state | | Descriptor: | (4S)-2-methyl-4-phenyl-3,4-dihydro-1H-isoquinolin-8-amine, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8Y90
 
 | | Structure of NET-Nefopam in outward-open state | | Descriptor: | (1S)-5-methyl-1-phenyl-1,3,4,6-tetrahydro-2,5-benzoxazocine, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8Y93
 
 | | Structure of NET-Amitriptyline in outward-open state | | Descriptor: | Amitriptyline, CHLORIDE ION, SODIUM ION, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
8Y94
 
 | |
8Y8Z
 
 | | Structure of NET-Maprotiline in outward-open state | | Descriptor: | CHLORIDE ION, SODIUM ION, Sodium-dependent noradrenaline transporter, ... | | Authors: | Zhang, H, Xu, E.H, Jiang, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Dimerization and antidepressant recognition at noradrenaline transporter.
Nature, 630, 2024
|
|
